Implementation of oscillations of neutrinos in matter in a framework with a Taylor expansion. More...
#include <PMNS_Maltoni.h>
Public Member Functions | |
| PMNS_Maltoni (int numNus) | |
| Constructor. More... | |
| virtual void | SetPremModel (OscProb::PremModel &prem) |
| virtual double | AvgProb (int flvi, int flvf, double E, double dE) |
| virtual double | AvgProb (int flvi, int flvf, double E, double cosT, double dcosT) |
| virtual double | AvgProb (int flvi, int flvf, double E, double dE, double cosT, double dcosT) |
| virtual double | AvgProbLoE (int flvi, int flvf, double LoE, double dLoE) |
| virtual double | AvgProbLoE (int flvi, int flvf, double LoE, double dLoE, double cosT, double dcosT) |
| virtual vectorD | AvgProbVector (int flvi, double E, double dE) |
| virtual vectorD | AvgProbVectorLoE (int flvi, double LoE, double dLoE) |
| virtual matrixD | AvgProbMatrix (int nflvi, int nflvf, double E, double dE) |
| virtual matrixD | AvgProbMatrixLoE (int nflvi, int nflvf, double LoE, double dLoE) |
| virtual double | ExtrapolationProb (int flvi, int flvf, double E, double dE) |
| Compute the probability of flvi going to flvf for an energy E+dE. More... | |
| virtual double | ExtrapolationProbLoE (int flvi, int flvf, double LoE, double dLoE) |
| Compute the probability of flvi going to flvf at LoE+dLoE. More... | |
| virtual double | ExtrapolationProbCosT (int flvi, int flvf, double cosT, double dcosT) |
| Compute the probability of flvi going to flvf for an angle cosT+dcosT. More... | |
| virtual void | SetIsOscProbAvg (bool isOscProbAvg) |
| Set flag for which averaging to use. More... | |
| virtual double | AvgProb (vectorC nu_in, int flvf, double E, double dE=0) |
| Compute the average probability over a bin of energy. More... | |
| virtual double | AvgProb (int flvi, int flvf, double E, double dE=0) |
| Compute the average probability over a bin of energy. More... | |
| virtual double | AvgProbLoE (vectorC nu_in, int flvf, double LoE, double dLoE=0) |
| Compute the average probability over a bin of L/E. More... | |
| virtual double | AvgProbLoE (int flvi, int flvf, double LoE, double dLoE=0) |
| Compute the average probability over a bin of L/E. More... | |
| virtual double | Prob (vectorC nu_in, int flvf) |
| Compute the probability of nu_in going to flvf. More... | |
| virtual double | Prob (vectorC nu_in, int flvf, double E) |
| virtual double | Prob (vectorC nu_in, int flvf, double E, double L) |
| virtual double | Prob (int flvi, int flvf) |
| Compute the probability of flvi going to flvf. More... | |
| virtual double | Prob (int flvi, int flvf, double E) |
| virtual double | Prob (int flvi, int flvf, double E, double L) |
| virtual vectorD | ProbVector (vectorC nu_in) |
| virtual vectorD | ProbVector (vectorC nu_in, double E) |
| flavours for energy E More... | |
| virtual vectorD | ProbVector (vectorC nu_in, double E, double L) |
| virtual vectorD | ProbVector (int flvi) |
| virtual vectorD | ProbVector (int flvi, double E) |
| virtual vectorD | ProbVector (int flvi, double E, double L) |
| virtual matrixD | ProbMatrix (int nflvi, int nflvf) |
| Compute the probability matrix. More... | |
| virtual matrixD | ProbMatrix (int nflvi, int nflvf, double E) |
| Compute the probability matrix for energy E. More... | |
| virtual matrixD | ProbMatrix (int nflvi, int nflvf, double E, double L) |
| virtual vectorD | AvgProbVector (vectorC nu_in, double E, double dE=0) |
| virtual vectorD | AvgProbVectorLoE (vectorC nu_in, double LoE, double dLoE=0) |
| Compute the average probability vector over a bin of L/E. More... | |
| virtual vectorC | GetMassEigenstate (int mi) |
| Get a neutrino mass eigenstate. More... | |
| virtual void | SetAngle (int i, int j, double th) |
| Set the mixing angle theta_ij. More... | |
| virtual void | SetDelta (int i, int j, double delta) |
| Set the CP phase delta_ij. More... | |
| virtual void | SetDm (int j, double dm) |
| Set the mass-splitting dm_j1 in eV^2. More... | |
| virtual double | GetAngle (int i, int j) |
| Get the mixing angle theta_ij. More... | |
| virtual double | GetDelta (int i, int j) |
| Get the CP phase delta_ij. More... | |
| virtual double | GetDm (int j) |
| Get the mass-splitting dm_j1 in eV^2. More... | |
| virtual double | GetDmEff (int j) |
| Get the effective mass-splitting dm_j1 in eV^2. More... | |
| virtual void | SetStdPars () |
| Set PDG 3-flavor parameters. More... | |
| virtual void | SetEnergy (double E) |
| Set the neutrino energy in GeV. More... | |
| virtual void | SetIsNuBar (bool isNuBar) |
| Set the anti-neutrino flag. More... | |
| virtual double | GetEnergy () |
| Get the neutrino energy in GeV. More... | |
| virtual bool | GetIsNuBar () |
| Get the anti-neutrino flag. More... | |
| virtual void | SetPath (NuPath p) |
| Set a single path. More... | |
| virtual void | SetPath (double length, double density, double zoa=0.5, int layer=0) |
| Set a single path. More... | |
| virtual void | SetPath (std::vector< NuPath > paths) |
| Set a path sequence. More... | |
| virtual void | AddPath (NuPath p) |
| Add a path to the sequence. More... | |
| virtual void | AddPath (double length, double density, double zoa=0.5, int layer=0) |
| Add a path to the sequence. More... | |
| virtual void | ClearPath () |
| Clear the path vector. More... | |
| virtual void | SetLength (double L) |
| Set a single path lentgh in km. More... | |
| virtual void | SetLength (vectorD L) |
| Set multiple path lengths. More... | |
| virtual void | SetDensity (double rho) |
| Set single path density in g/cm^3. More... | |
| virtual void | SetDensity (vectorD rho) |
| Set multiple path densities. More... | |
| virtual void | SetZoA (double zoa) |
| Set Z/A value for single path. More... | |
| virtual void | SetZoA (vectorD zoa) |
| Set multiple path Z/A values. More... | |
| virtual void | SetLayers (std::vector< int > lay) |
| Set multiple path layer indices. More... | |
| virtual void | SetStdPath () |
| Set standard neutrino path. More... | |
| virtual std::vector< NuPath > | GetPath () |
| Get the neutrino path sequence. More... | |
| virtual vectorD | GetSamplePoints (double LoE, double dLoE) |
| Compute the sample points for a bin of L/E with width dLoE. More... | |
| virtual void | SetUseCache (bool u=true) |
| Set caching on/off. More... | |
| virtual void | ClearCache () |
| Clear the cache. More... | |
| virtual void | SetMaxCache (int mc=1e6) |
| Set max cache size. More... | |
| virtual void | SetAvgProbPrec (double prec) |
| Set the AvgProb precision. More... | |
Protected Member Functions | |
| virtual vectorD | GetSamplePointsAvgClass (double LoE, double dLoE) |
| Compute the sample points fo a bin of L/E with width dLoE. More... | |
| virtual vectorD | GetSamplePointsAvgClass (double E, double cosT, double dcosT) |
| Compute the sample points for a bin of cosT with width dcosT. More... | |
| virtual matrixC | GetSamplePointsAvgClass (double InvE, double dInvE, double cosT, double dcosT) |
| virtual void | UpdateHam ()=0 |
| Build the full Hamiltonian. More... | |
| virtual void | InitializeTaylorsVectors () |
| Initialize all member vectors with zeros. More... | |
| virtual void | SetwidthBin (double dE, double dcosT) |
| Set bin's widths for energy and angle. More... | |
| virtual void | SetCosT (double cosT) |
| Set neutrino angle. More... | |
| virtual void | BuildKE (double L) |
| build K matrix for the inverse of energy in mass basis More... | |
| virtual void | BuildKcosT () |
| build K matrix for angle in flavor basis More... | |
| virtual double | LnDerivative () |
| Compute the derivation of one layer's length. More... | |
| virtual void | PropagatePathTaylor (NuPath p) |
| Propagate neutrino through a single path. More... | |
| virtual void | PropagateTaylor () |
| Propagate neutrino through full path. More... | |
| virtual void | rotateS () |
| Rotate the S matrix. More... | |
| virtual void | rotateK () |
| Rotate one K matrix. More... | |
| virtual void | MultiplicationRuleS () |
| Product between two S matrices. More... | |
| virtual void | MultiplicationRuleK (Eigen::MatrixXcd &K) |
| Product between two K matrices. More... | |
| void | SolveK (Eigen::MatrixXcd &K, vectorD &lambda, matrixC &V) |
| Solve the K matrix for eigenvectors and eigenvalues. More... | |
| template<typename T > | |
| void | TemplateSolver (Eigen::MatrixXcd &K, vectorD &lambda, matrixC &V) |
| Auxiliary function to choose eigensystem method. More... | |
| virtual double | AvgFormula (int flvi, int flvf, double dbin, vectorD flambda, matrixC fV) |
| Formula for the average probability over a bin of width dbin. More... | |
| virtual double | AvgFormulaExtrapolation (int flvi, int flvf, double dE, vectorD flambda, matrixC fV) |
| Formula for the extrapolation of probability. More... | |
| virtual double | AvgAlgo (int flvi, int flvf, double LoE, double dLoE, double L) |
| virtual double | AvgAlgo (int flvi, int flvf, double InvE, double dInvE, double cosT, double dcosT) |
| virtual double | AvgAlgoCosT (int flvi, int flvf, double E, double cosT, double dcosT) |
| virtual double | AlgorithmDensityMatrix (int flvi, int flvf) |
| Algorithm for the transformations on the density matrix. More... | |
| virtual void | RotateDensityM (bool to_basis, matrixC V) |
| Apply rotation to the density matrix. More... | |
| virtual void | HadamardProduct (vectorD lambda, double dbin) |
| Apply an Hadamard Product to the density matrix. More... | |
| virtual void | InitializeVectors () |
| virtual bool | TryCache () |
| Try to find a cached eigensystem. More... | |
| virtual void | FillCache () |
| Cache the current eigensystem. More... | |
| virtual void | SetCurPath (NuPath p) |
| Set the path currently in use by the class. More... | |
| virtual void | SetAtt (double att, int idx) |
| Set one of the path attributes. More... | |
| virtual void | SetAtt (vectorD att, int idx) |
| Set all values of a path attribute. More... | |
| virtual void | RotateH (int i, int j, matrixC &Ham) |
| Rotate the Hamiltonian by theta_ij and delta_ij. More... | |
| virtual void | RotateState (int i, int j) |
| Rotate the neutrino state by theta_ij and delta_ij. More... | |
| virtual void | BuildHms () |
| Build the matrix of masses squared. More... | |
| virtual void | SolveHam ()=0 |
| virtual void | ResetToFlavour (int flv) |
| Reset neutrino state to pure flavour flv. More... | |
| virtual void | SetPureState (vectorC nu_in) |
| Set the initial state from a pure state. More... | |
| virtual void | PropagatePath (NuPath p) |
| Propagate neutrino through a single path. More... | |
| virtual void | Propagate () |
| Propagate neutrino through full path. More... | |
| virtual double | P (int flv) |
| Return the probability of final state in flavour flv. More... | |
| virtual vectorD | GetProbVector () |
| virtual std::vector< int > | GetSortedIndices (const vectorD x) |
| Get indices that sort a vector. More... | |
| virtual vectorD | ConvertEtoLoE (double E, double dE) |
Protected Attributes | |
| double | fcosT |
| Cosine of zenith angle. More... | |
| double | fdInvE |
| Bin's width for the inverse of energy. More... | |
| double | fdcosT |
| Bin's width for zenith angle. More... | |
| vectorD | flambdaInvE |
| Eigenvectors of K_invE. More... | |
| vectorD | flambdaCosT |
| Eigenvectors of K_cosTheta. More... | |
| matrixC | fVInvE |
| Eigenvalues of K_invE. More... | |
| Eigen::MatrixXcd | fKInvE |
| K matrix for the inverse of energy for the entire path. More... | |
| Eigen::MatrixXcd | fKcosT |
| K matrix for neutrino angle for the entire path. More... | |
| matrixC | fevolutionMatrixS |
| matrixC | fVcosT |
| Eigenvalues of K_cosTheta. More... | |
| matrixC | fSflavor |
| S matrix for one layer. More... | |
| matrixC | fKmass |
| K matrix in mass basis for one layer. More... | |
| matrixC | fKflavor |
| K matrix in flavor basis for one layer. More... | |
| matrixC | fdensityMatrix |
| The neutrino density matrix state. More... | |
| int | fLayer |
| Layer index. More... | |
| int | fdl |
| Length derivative. More... | |
| double | fDetRadius |
| Detector radius. More... | |
| double | fminRsq |
| Minimum square radius. More... | |
| OscProb::PremModel | fPrem |
| Earth model used. More... | |
| Eigen::MatrixXcd | fHam |
| The full Hamiltonian. More... | |
| bool | fIsOscProbAvg |
| Flag to call OscProb default or Maltoni average. More... | |
| int | fNumNus |
| Number of neutrino flavours. More... | |
| vectorD | fDm |
| m^2_i - m^2_1 in vacuum More... | |
| matrixD | fTheta |
| theta[i][j] mixing angle More... | |
| matrixD | fDelta |
| delta[i][j] CP violating phase More... | |
| vectorC | fNuState |
| The neutrino current state. More... | |
| matrixC | fHms |
| matrix H*2E in eV^2 More... | |
| vectorC | fPhases |
| Buffer for oscillation phases. More... | |
| vectorC | fBuffer |
| Buffer for neutrino state tranformations. More... | |
| vectorD | fEval |
| Eigenvalues of the Hamiltonian. More... | |
| matrixC | fEvec |
| Eigenvectors of the Hamiltonian. More... | |
| double | fEnergy |
| Neutrino energy. More... | |
| bool | fIsNuBar |
| Anti-neutrino flag. More... | |
| std::vector< NuPath > | fNuPaths |
| Vector of neutrino paths. More... | |
| NuPath | fPath |
| Current neutrino path. More... | |
| bool | fBuiltHms |
| Tag to avoid rebuilding Hms. More... | |
| bool | fGotES |
| Tag to avoid recalculating eigensystem. More... | |
| bool | fUseCache |
| Flag for whether to use caching. More... | |
| double | fCachePrec |
| Precision of cache matching. More... | |
| int | fMaxCache |
| Maximum cache size. More... | |
| double | fAvgProbPrec |
| AvgProb precision. More... | |
| std::unordered_set< EigenPoint > | fMixCache |
| Caching set of eigensystems. More... | |
| EigenPoint | fProbe |
| EigenpPoint to try. More... | |
Static Protected Attributes | |
| static const complexD | zero |
| zero in complex More... | |
| static const complexD | one |
| one in complex More... | |
| static const double | kKm2eV = 1.0 / 1.973269788e-10 |
| km to eV^-1 More... | |
| static const double | kK2 |
| mol/GeV^2/cm^3 to eV More... | |
| static const double | kGeV2eV = 1.0e+09 |
| GeV to eV. More... | |
| static const double | kNA = 6.022140857e23 |
| Avogadro constant. More... | |
| static const double | kGf = 1.1663787e-05 |
| G_F in units of GeV^-2. More... | |
Detailed Description
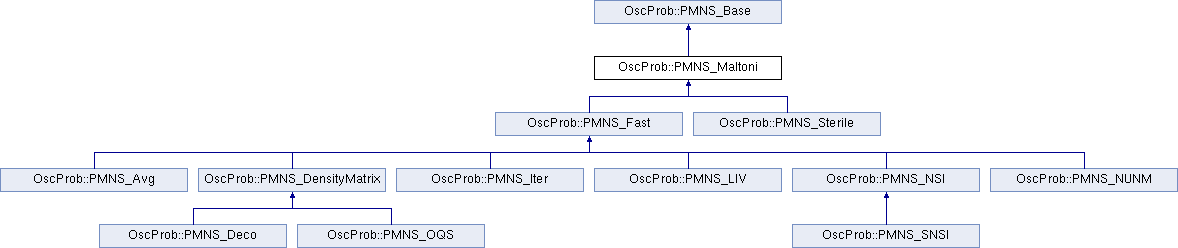
This class expands the PMNS_Base class including the use of a Taylor expansion to calculate the average on bins faster.
The model assumes a first order expansion over energy or direction for both dynamical variables at the same time or for only one.
This version is limited to classes where the vacuum Hamiltonian is inversely proportional to energy and the matter potential is independent of energy.
Reference: https://doi.org/10.48550/arXiv.2308.00037
- See also
- PMNS_Base
Definition at line 34 of file PMNS_Maltoni.h.
Constructor & Destructor Documentation
◆ PMNS_Maltoni()
| PMNS_Maltoni::PMNS_Maltoni | ( | int | numNus | ) |
Constructor.
- See also
- PMNS_Base::PMNS_Base
Definition at line 23 of file PMNS_Maltoni.cxx.
References fPrem, InitializeTaylorsVectors(), OscProb::PremModel::LoadModel(), OscProb::PMNS_Base::SetAvgProbPrec(), SetIsOscProbAvg(), and SetwidthBin().
Member Function Documentation
◆ AddPath() [1/2]
|
virtualinherited |
Add a path to the sequence defining attributes directly.
- Parameters
-
length - The length of the path segment in km density - The density of the path segment in g/cm^3 zoa - The effective Z/A of the path segment layer - An index to identify the layer type (e.g. earth inner core)
Definition at line 317 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::AddPath().
◆ AddPath() [2/2]
|
virtualinherited |
Add a path to the sequence.
- Parameters
-
p - A neutrino path segment
Definition at line 307 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fNuPaths.
Referenced by OscProb::PMNS_Base::AddPath(), OscProb::PMNS_Base::SetAtt(), OscProb::PMNS_Base::SetPath(), and SetTestPath().
◆ AlgorithmDensityMatrix()
|
protectedvirtual |
Algorithm for the transformations on the density matrix
- Parameters
-
flvi - The neutrino starting flavour. flvf - The neutrino final flavour.
- Returns
- Average neutrino oscillation probability
Definition at line 1099 of file PMNS_Maltoni.cxx.
References fdcosT, fdensityMatrix, fdInvE, fevolutionMatrixS, flambdaCosT, flambdaInvE, fVcosT, fVInvE, HadamardProduct(), OscProb::PMNS_Base::kGeV2eV, and RotateDensityM().
Referenced by AvgAlgo().
◆ AvgAlgo() [1/2]
|
protectedvirtual |
Algorithm for the compute of the average probability over a bin of 1oE and cosT
Algorithm for computing the average probability over a bin of 1oE and cosT
- Parameters
-
flvi - The neutrino starting flavour. flvf - The neutrino final flavour. InvE - The neutrino 1/E value in the bin center in GeV-1 dInvE - The 1/E bin width in GeV-1 cosT - The cosine of the neutrino angle dcosT - The cosT bin width
- Returns
- Average neutrino oscillation probability
Definition at line 1065 of file PMNS_Maltoni.cxx.
References AlgorithmDensityMatrix(), fDetRadius, OscProb::PremModel::FillPath(), fKcosT, fKInvE, flambdaCosT, flambdaInvE, fminRsq, fPrem, fVcosT, fVInvE, OscProb::EarthModelBase::GetNuPath(), InitializeTaylorsVectors(), PropagateTaylor(), SetCosT(), OscProb::PMNS_Base::SetEnergy(), OscProb::PMNS_Base::SetPath(), SetwidthBin(), and SolveK().
◆ AvgAlgo() [2/2]
|
protectedvirtual |
Algorithm for the compute of the average probability over a bin of LoE
Algorithm for the compute of the average probability over a bin of LoE
- Parameters
-
flvi - The neutrino starting flavour. flvf - The neutrino final flavour. LoE - The neutrino L/E value in the bin center in km/GeV dLoE - The L/E bin width in km/GeV L - The length of the path in km
- Returns
- Average neutrino oscillation probability
Definition at line 594 of file PMNS_Maltoni.cxx.
References AvgFormula(), fKInvE, flambdaInvE, fVInvE, InitializeTaylorsVectors(), OscProb::PMNS_Base::kGeV2eV, PropagateTaylor(), OscProb::PMNS_Base::SetEnergy(), SetwidthBin(), and SolveK().
Referenced by AvgProbLoE(), AvgProbMatrixLoE(), and AvgProbVectorLoE().
◆ AvgAlgoCosT()
|
protectedvirtual |
Algorithm for the compute of the average probability over a bin of cosT
Algorithm for the compute of the average probability over a bin of cosT
- Parameters
-
flvi - The neutrino starting flavour. flvf - The neutrino final flavour. E - The neutrino energy in GeV cosT - The cosine of the neutrino angle dcosT - The cosT bin width
- Returns
- Average neutrino oscillation probability
Definition at line 913 of file PMNS_Maltoni.cxx.
References AvgFormula(), fdcosT, fDetRadius, OscProb::PremModel::FillPath(), fKcosT, flambdaCosT, fminRsq, fPrem, fVcosT, OscProb::EarthModelBase::GetNuPath(), InitializeTaylorsVectors(), PropagateTaylor(), SetCosT(), OscProb::PMNS_Base::SetEnergy(), OscProb::PMNS_Base::SetPath(), SetwidthBin(), and SolveK().
◆ AvgFormula()
|
protectedvirtual |
Formula for the average probability of flvi going to flvf over a bin of width dbin using the Maltoni method.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
flvi - The neutrino starting flavour. flvf - The neutrino final flavour. dbin - The width of the bin lambda - The eigenvalues of K V - The eigenvectors of K
- Returns
- Average neutrino oscillation probability
Definition at line 450 of file PMNS_Maltoni.cxx.
References fevolutionMatrixS, OscProb::PMNS_Base::fNumNus, and OscProb::PMNS_Base::P().
Referenced by AvgAlgo(), AvgAlgoCosT(), AvgProb(), AvgProbMatrixLoE(), and AvgProbVectorLoE().
◆ AvgFormulaExtrapolation()
|
protectedvirtual |
Fomula for the propability for flvi going to flvf for an energy E+dE using the Maltoni method from a reference energy E.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
flvi - The neutrino starting flavour. flvf - The neutrino final flavour. dE - The energy variation in GeV lambda - The eigenvalues of K V - The eigenvectors of K
- Returns
- Neutrino oscillation probability
Definition at line 1289 of file PMNS_Maltoni.cxx.
References fevolutionMatrixS, OscProb::PMNS_Base::fNumNus, and OscProb::PMNS_Base::P().
Referenced by ExtrapolationProb(), ExtrapolationProbCosT(), and ExtrapolationProbLoE().
◆ AvgProb() [1/5]
|
virtual |
Compute the average probability over a bin of cosTheta with a Taylor expansion
Compute the average probability of flvi going to flvf over a bin of angle cost with width dcosT using the Maltoni method.
IMPORTANT: The PremModel object used must be set by this class using the function SetPremModel.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
flvi - The neutrino starting flavour. flvf - The neutrino final flavour. E - The neutrino energy in GeV cosT - The cosine of the neutrino angle dcosT - The cosT bin width
- Returns
- Average neutrino oscillation probability
Definition at line 876 of file PMNS_Maltoni.cxx.
References AvgFormula(), fdcosT, fDetRadius, OscProb::PremModel::FillPath(), fKcosT, flambdaCosT, fminRsq, fPrem, fVcosT, OscProb::EarthModelBase::GetNuPath(), InitializeTaylorsVectors(), PropagateTaylor(), SetCosT(), OscProb::PMNS_Base::SetEnergy(), OscProb::PMNS_Base::SetPath(), SetwidthBin(), and SolveK().
◆ AvgProb() [2/5]
|
virtual |
Compute the average probability over a bin of energy with a Taylor expansion
Compute the average probability of flvi going to flvf over a bin of energy E with width dE using the Maltoni method.
This gets transformed into L/E, since the oscillation terms have arguments linear in 1/E and not E.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
flvi - The neutrino starting flavour. flvf - The neutrino final flavour. E - The neutrino energy in GeV dE - The energy bin width in GeV
- Returns
- Average neutrino oscillation probability
Reimplemented from OscProb::PMNS_Base.
Definition at line 507 of file PMNS_Maltoni.cxx.
References OscProb::PMNS_Base::AvgProb(), AvgProbLoE(), OscProb::PMNS_Base::ConvertEtoLoE(), fIsOscProbAvg, OscProb::PMNS_Base::fNuPaths, and OscProb::PMNS_Base::Prob().
Referenced by AvgProb(), and AvgProbLoE().
◆ AvgProb() [3/5]
|
virtual |
Compute the average probabilit over a bin of cosTheta and energy with a Taylor expansion
Compute the average probability of flvi going to flvf over a bin of energy E and angle cosT with width dE and dcosT using the Maltoni method.
IMPORTANT: The PremModel object used must be set by this class using the function SetPremModel.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
flvi - The neutrino starting flavour. flvf - The neutrino final flavour. E - The neutrino energy in GeV dE - The energy bin width in GeV cosT - The cosine of the neutrino angle dcosT - The cosT bin width
- Returns
- Average neutrino oscillation probability
Definition at line 961 of file PMNS_Maltoni.cxx.
References AvgProb(), AvgProbLoE(), OscProb::PMNS_Base::ConvertEtoLoE(), OscProb::PMNS_Base::fNuPaths, and OscProb::PMNS_Base::Prob().
◆ AvgProb() [4/5]
|
virtual |
Compute the average probability of flvi going to flvf over a bin of energy E with width dE.
This gets transformed into L/E, since the oscillation terms have arguments linear in L/E and not E.
This function works best for single paths. In multiple paths the accuracy may be somewhat worse. If needed, average over smaller energy ranges.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
flvi - The neutrino starting flavour. flvf - The neutrino final flavour. E - The neutrino energy in the bin center in GeV dE - The energy bin width in GeV
- Returns
- Average neutrino oscillation probability
Reimplemented from OscProb::PMNS_Base.
Definition at line 98 of file PMNS_Base.cxx.
◆ AvgProb() [5/5]
|
virtual |
Compute the average probability of nu_in going to flvf over a bin of energy E with width dE.
This gets transformed into L/E, since the oscillation terms have arguments linear in L/E and not E.
This function works best for single paths. In multiple paths the accuracy may be somewhat worse. If needed, average over smaller energy ranges.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
nu_in - The neutrino initial state in flavour. flvf - The neutrino final flavour. E - The neutrino energy in the bin center in GeV dE - The energy bin width in GeV
- Returns
- Average neutrino oscillation probability
Reimplemented from OscProb::PMNS_Base.
Definition at line 88 of file PMNS_Base.cxx.
◆ AvgProbLoE() [1/4]
|
virtual |
Compute the average probability over a bin of LoE with a Taylor expansion
Compute the average probability of flvi going to flvf over a bin of energy L/E with width dLoE using the Maltoni method.
The probabilities are weighted by (L/E)^-2 so that event density is flat in energy. This avoids giving too much weight to low energies. Better approximations would be achieved if we used an interpolated event density.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
flvi - The neutrino starting flavour. flvf - The neutrino final flavour. LoE - The neutrino L/E value in the bin center in km/GeV dLoE - The L/E bin width in km/GeV
- Returns
- Average neutrino oscillation probability
Reimplemented from OscProb::PMNS_Base.
Definition at line 548 of file PMNS_Maltoni.cxx.
References AvgAlgo(), OscProb::PMNS_Base::AvgProbLoE(), fIsOscProbAvg, OscProb::PMNS_Base::fNuPaths, OscProb::PMNS_Base::fPath, GetSamplePointsAvgClass(), OscProb::NuPath::length, and OscProb::PMNS_Base::Prob().
Referenced by AvgProb(), and AvgProbLoE().
◆ AvgProbLoE() [2/4]
|
virtual |
Compute the average probability over a bin of cosTheta and LoE with a Taylor expansion
Compute the average probability of flvi going to flvf over a bin of energy L/E and cosT with width dLoE and dcosT using the Maltoni method.
The probabilities are weighted by (L/E)^-2 so that event density is flat in energy. This avoids giving too much weight to low energies. Better approximations would be achieved if we used an interpolated event density.
IMPORTANT: The PremModel object used must be set by this class using the function SetPremModel.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
flvi - The neutrino starting flavour. flvf - The neutrino final flavour. LoE - The neutrino L/E value in the bin center in km/GeV dLoE - The L/E bin width in km/GeV cosT - The cosine of the neutrino angle dcosT - The cosT bin width
- Returns
- Average neutrino oscillation probability
chg ici
Definition at line 1008 of file PMNS_Maltoni.cxx.
References AvgAlgo(), AvgProb(), AvgProbLoE(), OscProb::PMNS_Base::fPath, GetSamplePointsAvgClass(), OscProb::NuPath::length, and OscProb::PMNS_Base::Prob().
◆ AvgProbLoE() [3/4]
|
virtual |
Compute the average probability of flvi going to flvf over a bin of L/E with width dLoE.
The probabilities are weighted by (L/E)^-2 so that event density is flat in energy. This avoids giving too much weight to low energies. Better approximations would be achieved if we used an interpolated event density.
This function works best for single paths. In multiple paths the accuracy may be somewhat worse. If needed, average over smaller L/E ranges.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
flvi - The neutrino starting flavour. flvf - The neutrino final flavour. LoE - The neutrino L/E value in the bin center in km/GeV dLoE - The L/E bin width in km/GeV
- Returns
- Average neutrino oscillation probability
Reimplemented from OscProb::PMNS_Base.
Definition at line 102 of file PMNS_Base.cxx.
◆ AvgProbLoE() [4/4]
|
virtual |
Compute the average probability of nu_in going to flvf over a bin of L/E with width dLoE.
The probabilities are weighted by (L/E)^-2 so that event density is flat in energy. This avoids giving too much weight to low energies. Better approximations would be achieved if we used an interpolated event density.
This function works best for single paths. In multiple paths the accuracy may be somewhat worse. If needed, average over smaller L/E ranges.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
nu_in - The neutrino intial state in flavour basis. flvf - The neutrino final flavour. LoE - The neutrino L/E value in the bin center in km/GeV dLoE - The L/E bin width in km/GeV
- Returns
- Average neutrino oscillation probability
Reimplemented from OscProb::PMNS_Base.
Definition at line 92 of file PMNS_Base.cxx.
◆ AvgProbMatrix()
|
virtual |
Compute the average probability matrix over a bin of energy using a Taylor expansion
Compute the average probability matrix for nflvi and nflvf over a bin of energy E with width dE using the Maltoni method.
- Parameters
-
nflvi - The number of initial flavours in the matrix. nflvf - The number of final flavours in the matrix. E - The neutrino energy in the bin center in GeV dE - The energy bin width in GeV
- Returns
- Average neutrino oscillation probabilities
Reimplemented from OscProb::PMNS_Base.
Definition at line 767 of file PMNS_Maltoni.cxx.
References OscProb::PMNS_Base::AvgProbMatrix(), AvgProbMatrixLoE(), OscProb::PMNS_Base::ConvertEtoLoE(), fIsOscProbAvg, OscProb::PMNS_Base::fNuPaths, and OscProb::PMNS_Base::ProbMatrix().
◆ AvgProbMatrixLoE()
|
virtual |
Compute the average probability matrix over a bin of L/E using a Taylor expansion
Compute the average probability matrix for nflvi and nflvf over a bin of L/E with width dLoE using the Maltoni method.
- Parameters
-
nflvi - The number of initial flavours in the matrix. nflvf - The number of final flavours in the matrix. LoE - The neutrino L/E value in the bin center in km/GeV dLoE - The L/E bin width in km/GeV
- Returns
- Average neutrino oscillation probabilities
Reimplemented from OscProb::PMNS_Base.
Definition at line 800 of file PMNS_Maltoni.cxx.
References AvgAlgo(), AvgFormula(), OscProb::PMNS_Base::AvgProbMatrixLoE(), fdInvE, fIsOscProbAvg, flambdaInvE, OscProb::PMNS_Base::fNuPaths, OscProb::PMNS_Base::fPath, fVInvE, GetSamplePointsAvgClass(), OscProb::PMNS_Base::kGeV2eV, OscProb::NuPath::length, and OscProb::PMNS_Base::ProbMatrix().
Referenced by AvgProbMatrix().
◆ AvgProbVector() [1/2]
|
virtual |
Compute the average probability vector over a bin of energy using a Taylor expansion
Compute the average probability of flvi going to any flavour over a bin of energy E with width dE using the Maltoni method.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
flvi - The neutrino starting flavour. E - The neutrino energy in GeV dE - The energy bin width in GeV
- Returns
- Average probability for all outgoing flavours
Reimplemented from OscProb::PMNS_Base.
Definition at line 671 of file PMNS_Maltoni.cxx.
References OscProb::PMNS_Base::AvgProbVector(), AvgProbVectorLoE(), OscProb::PMNS_Base::ConvertEtoLoE(), fIsOscProbAvg, OscProb::PMNS_Base::fNumNus, OscProb::PMNS_Base::fNuPaths, and OscProb::PMNS_Base::ProbVector().
◆ AvgProbVector() [2/2]
Compute the average probability vector over a bin of energy
Compute the average probability of nu_in going to all flavours over a bin of energy E with width dE.
This gets transformed into L/E, since the oscillation terms have arguments linear in L/E and not E.
This function works best for single paths. In multiple paths the accuracy may be somewhat worse. If needed, average over smaller energy ranges.
- Parameters
-
nu_in - The neutrino initial state in flavour. E - The neutrino energy in the bin center in GeV dE - The energy bin width in GeV
- Returns
- Average neutrino oscillation probabilities
Definition at line 1753 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::AvgProbVectorLoE(), OscProb::PMNS_Base::ConvertEtoLoE(), OscProb::PMNS_Base::fNumNus, OscProb::PMNS_Base::fNuPaths, and OscProb::PMNS_Base::ProbVector().
Referenced by AvgProbVector(), and OscProb::PMNS_Base::AvgProbVector().
◆ AvgProbVectorLoE() [1/2]
|
virtual |
Compute the average probability vector over a bin of L/E using a Taylor expansion
Compute the average probability of flvi going to any flavour over a bin of L/E with width dLoE using the Maltoni method.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
flvi - The neutrino starting flavour. LoE - The neutrino L/E value in the bin center in km/GeV dLoE - The L/E bin width in km/GeV
- Returns
- Average probability for all outgoing flavours
Reimplemented from OscProb::PMNS_Base.
Definition at line 708 of file PMNS_Maltoni.cxx.
References AvgAlgo(), AvgFormula(), OscProb::PMNS_Base::AvgProbVectorLoE(), fdInvE, fIsOscProbAvg, flambdaInvE, OscProb::PMNS_Base::fNumNus, OscProb::PMNS_Base::fNuPaths, OscProb::PMNS_Base::fPath, fVInvE, GetSamplePointsAvgClass(), OscProb::PMNS_Base::kGeV2eV, OscProb::NuPath::length, and OscProb::PMNS_Base::ProbVector().
Referenced by AvgProbVector().
◆ AvgProbVectorLoE() [2/2]
Compute the average probability of nu_in going to all flavours over a bin of L/E with width dLoE.
The probabilities are weighted by (L/E)^-2 so that event density is flat in energy. This avoids giving too much weight to low energies. Better approximations would be achieved if we used an interpolated event density.
This function works best for single paths. In multiple paths the accuracy may be somewhat worse. If needed, average over smaller L/E ranges.
- Parameters
-
nu_in - The neutrino intial state in flavour basis. LoE - The neutrino L/E value in the bin center in km/GeV dLoE - The L/E bin width in km/GeV
- Returns
- Average neutrino oscillation probabilities
Definition at line 1791 of file PMNS_Base.cxx.
References OscProb::AvgPath(), OscProb::PMNS_Base::fNumNus, OscProb::PMNS_Base::fNuPaths, OscProb::PMNS_Base::fPath, OscProb::PMNS_Base::GetSamplePoints(), OscProb::NuPath::length, OscProb::PMNS_Base::ProbVector(), OscProb::PMNS_Base::SetCurPath(), and OscProb::PMNS_Base::SetEnergy().
Referenced by OscProb::PMNS_Base::AvgProbVector(), AvgProbVectorLoE(), and OscProb::PMNS_Base::AvgProbVectorLoE().
◆ BuildHms()
|
protectedvirtualinherited |
Build Hms = H*2E, where H is the Hamiltonian in vacuum on flavour basis and E is the neutrino energy in eV. Hms is effectively the matrix of masses squared.
This is a hermitian matrix, so only the upper triangular part needs to be filled
The construction of the Hamiltonian avoids computing terms that are simply zero. This has a big impact in the computation time.
Reimplemented in OscProb::PMNS_Decay, OscProb::PMNS_OQS, and OscProb::PMNS_SNSI.
Definition at line 955 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::ClearCache(), OscProb::PMNS_Base::fBuiltHms, OscProb::PMNS_Base::fDm, OscProb::PMNS_Base::fGotES, OscProb::PMNS_Base::fHms, OscProb::PMNS_Base::fNumNus, and OscProb::PMNS_Base::RotateH().
Referenced by OscProb::PMNS_OQS::BuildHms(), OscProb::PMNS_Sterile::SolveHam(), and OscProb::PMNS_Fast::SolveHamMatter().
◆ BuildKcosT()
|
protectedvirtual |
Build K matrix for the zenith angle in flavor basis.
Definition at line 111 of file PMNS_Maltoni.cxx.
References fHam, fKflavor, OscProb::PMNS_Base::fNumNus, OscProb::PMNS_Base::kKm2eV, LnDerivative(), and UpdateHam().
Referenced by PropagatePathTaylor().
◆ BuildKE()
|
protectedvirtual |
Build K matrix for the inverse of energy in mass basis.
The variable for which a Taylor expansion is done here is not directly the energy but the inverse of it. This change of variable allow to express the hamiltonien as linear with respect to this new variable.
- Parameters
-
L - The length of the layer in km
Definition at line 136 of file PMNS_Maltoni.cxx.
References OscProb::PMNS_Base::fEval, OscProb::PMNS_Base::fEvec, OscProb::PMNS_Base::fHms, OscProb::PMNS_Base::fIsNuBar, fKmass, OscProb::PMNS_Base::fNumNus, and OscProb::PMNS_Base::kKm2eV.
Referenced by PropagatePathTaylor().
◆ ClearCache()
|
virtualinherited |
Clear the cache
Definition at line 111 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fMixCache.
Referenced by OscProb::PMNS_Base::BuildHms(), OscProb::PMNS_Base::PMNS_Base(), OscProb::PMNS_NUNM::SetAlpha(), OscProb::PMNS_LIV::SetaT(), OscProb::PMNS_NSI::SetCoupByIndex(), OscProb::PMNS_LIV::SetcT(), OscProb::PMNS_NSI::SetEps(), and OscProb::PMNS_NUNM::SetFracVnc().
◆ ClearPath()
|
virtualinherited |
Clear the path vector.
Definition at line 287 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fNuPaths.
Referenced by OscProb::PMNS_Base::SetAtt(), and OscProb::PMNS_Base::SetPath().
◆ ConvertEtoLoE()
|
protectedvirtualinherited |
Convert a bin of energy into a bin of L/E
- Parameters
-
E - energy bin center in GeV dE - energy bin width in GeV
- Returns
- The L/E bin center and width in km/GeV
Definition at line 1516 of file PMNS_Base.cxx.
References OscProb::AvgPath(), OscProb::PMNS_Base::fNuPaths, OscProb::PMNS_Base::fPath, OscProb::NuPath::length, and OscProb::PMNS_Base::SetCurPath().
Referenced by AvgProb(), OscProb::PMNS_Base::AvgProb(), AvgProbMatrix(), OscProb::PMNS_Base::AvgProbMatrix(), AvgProbVector(), and OscProb::PMNS_Base::AvgProbVector().
◆ ExtrapolationProb()
|
virtual |
Compute the propability for flvi going to flvf for an energy E+dE using the Maltoni method from a reference energy E.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
flvi - The neutrino starting flavour. flvf - The neutrino final flavour. E - The reference energy in GeV dE - The energy variation in GeV
- Returns
- Neutrino oscillation probability
Definition at line 1341 of file PMNS_Maltoni.cxx.
References AvgFormulaExtrapolation(), fdInvE, fKInvE, flambdaInvE, fVInvE, InitializeTaylorsVectors(), OscProb::PMNS_Base::kGeV2eV, PropagateTaylor(), OscProb::PMNS_Base::SetEnergy(), SetwidthBin(), and SolveK().
◆ ExtrapolationProbCosT()
|
virtual |
Compute the propability for flvi going to flvf for an angle cosT+dcosT using the Maltoni method from a reference angle cosT.
IMPORTANT: The PremModel object used must be set by this class using the function SetPremModel.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
flvi - The neutrino starting flavour. flvf - The neutrino final flavour. cosT - The reference angle dcosT - The angle variation
- Returns
- Neutrino oscillation probability
Definition at line 1420 of file PMNS_Maltoni.cxx.
References AvgFormulaExtrapolation(), fdcosT, fDetRadius, OscProb::PremModel::FillPath(), fKcosT, flambdaCosT, fminRsq, fPrem, fVcosT, OscProb::EarthModelBase::GetNuPath(), InitializeTaylorsVectors(), PropagateTaylor(), SetCosT(), OscProb::PMNS_Base::SetPath(), SetwidthBin(), and SolveK().
◆ ExtrapolationProbLoE()
|
virtual |
Compute the propability for flvi going to flvf for an energy LoE+dLoE using the Maltoni method from a reference value LoE.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
flvi - The neutrino starting flavour. flvf - The neutrino final flavour. LoE - The reference energy in GeV dLoE - The energy variation in GeV
- Returns
- Neutrino oscillation probability
Definition at line 1377 of file PMNS_Maltoni.cxx.
References AvgFormulaExtrapolation(), OscProb::AvgPath(), fKInvE, flambdaInvE, OscProb::PMNS_Base::fNuPaths, OscProb::PMNS_Base::fPath, fVInvE, InitializeTaylorsVectors(), OscProb::PMNS_Base::kGeV2eV, OscProb::NuPath::length, PropagateTaylor(), OscProb::PMNS_Base::SetCurPath(), OscProb::PMNS_Base::SetEnergy(), SetwidthBin(), and SolveK().
◆ FillCache()
|
protectedvirtualinherited |
If using caching, save the eigensystem in memory
Reimplemented in OscProb::PMNS_LIV, and OscProb::PMNS_SNSI.
Definition at line 157 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fEnergy, OscProb::EigenPoint::fEval, OscProb::PMNS_Base::fEval, OscProb::EigenPoint::fEvec, OscProb::PMNS_Base::fEvec, OscProb::PMNS_Base::fIsNuBar, OscProb::PMNS_Base::fMaxCache, OscProb::PMNS_Base::fMixCache, OscProb::PMNS_Base::fNumNus, OscProb::PMNS_Base::fPath, OscProb::PMNS_Base::fProbe, OscProb::PMNS_Base::fUseCache, and OscProb::EigenPoint::SetVars().
Referenced by OscProb::PMNS_Sterile::SolveHam(), and OscProb::PMNS_Fast::SolveHamMatter().
◆ GetAngle()
|
virtualinherited |
Get the mixing angle theta_ij in radians.
Requires that i<j. Will notify you if input is wrong. If i>j, will assume reverse order and swap i and j.
- Parameters
-
i,j - the indices of theta_ij
Definition at line 570 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fNumNus, and OscProb::PMNS_Base::fTheta.
◆ GetDelta()
|
virtualinherited |
Get the CP phase delta_ij in radians.
Requires that i+1<j. Will notify you if input is wrong. If i>j, will assume reverse order and swap i and j.
- Parameters
-
i,j - the indices of delta_ij
Definition at line 638 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fDelta, and OscProb::PMNS_Base::fNumNus.
◆ GetDm()
|
virtualinherited |
Get the mass-splitting dm_j1 = (m_j^2 - m_1^2) in eV^2
Requires that j>1. Will notify you if input is wrong.
- Parameters
-
j - the index of dm_j1
Definition at line 696 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fDm, and OscProb::PMNS_Base::fNumNus.
◆ GetDmEff()
|
virtualinherited |
Get the effective mass-splitting dm_j1 in matter in eV^2
Requires that j>1. Will notify you if input is wrong.
- Parameters
-
j - the index of dm_j1
Definition at line 732 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fDm, OscProb::PMNS_Base::fEnergy, OscProb::PMNS_Base::fEval, OscProb::PMNS_Base::fNumNus, OscProb::PMNS_Base::GetSortedIndices(), OscProb::PMNS_Base::kGeV2eV, and OscProb::PMNS_Base::SolveHam().
◆ GetEnergy()
|
virtualinherited |
Get the neutrino energy in GeV.
Definition at line 255 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fEnergy.
◆ GetIsNuBar()
|
virtualinherited |
Get the anti-neutrino flag.
Definition at line 261 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fIsNuBar.
◆ GetMassEigenstate()
|
virtualinherited |
Get the neutrino mass eigenstate in vacuum
States are:
0 = m_1, 1 = m_2, 2 = m_3, etc.
- Parameters
-
mi - the mass eigenstate index
- Returns
- The mass eigenstate
Definition at line 795 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fNumNus, OscProb::PMNS_Base::fNuState, OscProb::PMNS_Base::ResetToFlavour(), and OscProb::PMNS_Base::RotateState().
◆ GetPath()
|
virtualinherited |
Get the vector of neutrino paths.
Definition at line 300 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fNuPaths.
◆ GetProbVector()
|
protectedvirtualinherited |
Return vector of probabilities from final state
Get the vector of probabilities for current state
- Returns
- Neutrino oscillation probabilities
Definition at line 1233 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fNumNus, and OscProb::PMNS_Base::P().
Referenced by OscProb::PMNS_Base::ProbVector().
◆ GetSamplePoints()
|
virtualinherited |
Compute the sample points for a bin of L/E with width dLoE
This is used for averaging the probability over a bin of L/E. It should be a private function, but I'm keeping it public for now for debugging purposes. The number of sample points seems too high for most purposes. The number of subdivisions needs to be optimized.
- Parameters
-
LoE - The neutrino L/E value in the bin center in km/GeV dLoE - The L/E bin width in km/GeV
Definition at line 1985 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fAvgProbPrec, OscProb::PMNS_Base::fEnergy, OscProb::PMNS_Base::fEval, OscProb::PMNS_Base::fNumNus, OscProb::PMNS_Base::kGeV2eV, OscProb::PMNS_Base::kKm2eV, and OscProb::PMNS_Base::SolveHam().
Referenced by OscProb::PMNS_Base::AvgProbLoE(), OscProb::PMNS_Base::AvgProbMatrixLoE(), and OscProb::PMNS_Base::AvgProbVectorLoE().
◆ GetSamplePointsAvgClass() [1/3]
|
protectedvirtual |
Compute the sample points for a bin of cosTheta with width dcosTheta
This is used to increase the precision of the average probability over a bin of cosT, calculated using the Maltoni method.
- Parameters
-
E - The neutrino Energy value GeV cosT - The neutrino cosT value in the bin center dcosT - The cosT bin width
- Returns
- Vector of sample cosT points
Definition at line 1243 of file PMNS_Maltoni.cxx.
References OscProb::PMNS_Base::fAvgProbPrec.
◆ GetSamplePointsAvgClass() [2/3]
|
protectedvirtual |
Compute the sample points for a bin of E and cosT with width dE and dcosT
Compute the sample points for a bin of 1oE and cosTheta with width d1oE and dcosTheta
This is used to increase the precision of the average probability over a bin of L/E and cosT, calculated using the Maltoni method.
- Parameters
-
InvE - The neutrino 1/E value in the bin center in GeV-1 dInvE - The 1/E bin width in GeV-1 cosT - The neutrino cosT value in the bin center dcosT - The cosT bin width
- Returns
- Matrix of sample InvE and cosT points
Definition at line 1131 of file PMNS_Maltoni.cxx.
References OscProb::PMNS_Base::fAvgProbPrec.
◆ GetSamplePointsAvgClass() [3/3]
|
protectedvirtual |
Compute the sample points for a bin of L/E with width dLoE.
This is used to increase the precision of the average probability over a bin of L/E, calculated using the Maltoni method.
- Parameters
-
LoE - The neutrino L/E value in the bin center in km/GeV dLoE - The L/E bin width in km/GeV
- Returns
- Vector of sample L/E points
Definition at line 628 of file PMNS_Maltoni.cxx.
References OscProb::PMNS_Base::fAvgProbPrec.
Referenced by AvgProbLoE(), AvgProbMatrixLoE(), and AvgProbVectorLoE().
◆ GetSortedIndices()
Get the indices of the sorted x vector
- Parameters
-
x - input vector
- Returns
- The vector of sorted indices
Definition at line 715 of file PMNS_Base.cxx.
Referenced by OscProb::PMNS_Base::GetDmEff().
◆ HadamardProduct()
|
protectedvirtual |
Apply an Hadamard Product to the density matrix
- Parameters
-
lambda - Eigenvalues of the K matrix dbin - Width of the bin
Definition at line 1209 of file PMNS_Maltoni.cxx.
References fdensityMatrix, and OscProb::PMNS_Base::fNumNus.
Referenced by AlgorithmDensityMatrix().
◆ InitializeTaylorsVectors()
|
protectedvirtual |
Set vector sizes and initialize elements to zero. Initialize diagonal elements of S to one
Definition at line 37 of file PMNS_Maltoni.cxx.
References fdensityMatrix, fDetRadius, fdl, fevolutionMatrixS, fKcosT, fKflavor, fKInvE, fKmass, flambdaCosT, flambdaInvE, fLayer, OscProb::PMNS_Base::fNumNus, fPrem, fSflavor, fVcosT, fVInvE, OscProb::PremModel::GetDetRadius(), and OscProb::PremModel::GetPremLayers().
Referenced by AvgAlgo(), AvgAlgoCosT(), AvgProb(), ExtrapolationProb(), ExtrapolationProbCosT(), ExtrapolationProbLoE(), and PMNS_Maltoni().
◆ InitializeVectors()
|
protectedvirtualinherited |
Initialize all member vectors with zeros
Set vector sizes and initialize elements to zero.
Definition at line 79 of file PMNS_Base.cxx.
Referenced by OscProb::PMNS_Base::PMNS_Base().
◆ LnDerivative()
|
protectedvirtual |
Compute the derivative of one layer's length depending on the angle
Definition at line 189 of file PMNS_Maltoni.cxx.
References fcosT, fDetRadius, fdl, fLayer, fminRsq, fPrem, and OscProb::PremModel::GetPremLayers().
Referenced by BuildKcosT().
◆ MultiplicationRuleK()
|
protectedvirtual |
Product between two K matrices.
This is used to calculate the matrix K corresponding to the propagation between the beginning of the path and the end of the current layer.
The matrix fKflavor correspond to the propagation between the beginning and the end of the layer.
- Parameters
-
K - The current K matrix
Definition at line 305 of file PMNS_Maltoni.cxx.
References fevolutionMatrixS, fKflavor, and OscProb::PMNS_Base::fNumNus.
Referenced by PropagatePathTaylor().
◆ MultiplicationRuleS()
|
protectedvirtual |
Product between two S matrices.
This is used to calculate the matrix S corresponding to the propagation between the beginning of the path and the end of the current layer.
The matrix fevolutionMatrixS represent the propagation between the beginning of the path and the beginning of the current layer. This matrix is updated after every layer with this function. The matrix fSflavor represent the propagation between the beginning and the end of the layer.
Definition at line 276 of file PMNS_Maltoni.cxx.
References fevolutionMatrixS, OscProb::PMNS_Base::fNumNus, and fSflavor.
Referenced by PropagatePathTaylor().
◆ P()
|
protectedvirtualinherited |
Compute oscillation probability of flavour flv from current state
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
flv - The neutrino final flavour.
- Returns
- Neutrino oscillation probability
Reimplemented in OscProb::PMNS_DensityMatrix.
Definition at line 1058 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fNumNus, and OscProb::PMNS_Base::fNuState.
Referenced by AvgFormula(), AvgFormulaExtrapolation(), OscProb::PMNS_Base::GetProbVector(), and OscProb::PMNS_Base::Prob().
◆ Prob() [1/6]
|
virtualinherited |
Compute the probability of flvi going to flvf.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
flvi - The neutrino starting flavour. flvf - The neutrino final flavour.
- Returns
- Neutrino oscillation probability
Definition at line 1091 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::P(), OscProb::PMNS_Base::Propagate(), and OscProb::PMNS_Base::ResetToFlavour().
◆ Prob() [2/6]
|
virtualinherited |
Compute the probability of flvi going to flvf for energy E
Compute the probability of flvi going to flvf for a given energy in GeV.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
flvi - The neutrino starting flavour. flvf - The neutrino final flavour. E - The neutrino energy in GeV
- Returns
- Neutrino oscillation probability
Definition at line 1160 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::Prob(), and OscProb::PMNS_Base::SetEnergy().
◆ Prob() [3/6]
|
virtualinherited |
Compute the probability of flvi going to flvf for energy E and distance L
Compute the probability of flvi going to flvf for a given energy in GeV and distance in km in a single path.
If the path sequence is not a single path, a new single path will be created and the previous sequence will be lost.
Don't use this if you want to propagate over multiple path segments.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
flvi - The neutrino starting flavour. flvf - The neutrino final flavour. E - The neutrino energy in GeV L - The neutrino path length in km
- Returns
- Neutrino oscillation probability
Definition at line 1219 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::Prob(), OscProb::PMNS_Base::SetEnergy(), and OscProb::PMNS_Base::SetLength().
◆ Prob() [4/6]
|
virtualinherited |
Compute the probability of nu_in going to flvf.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
nu_in - The neutrino initial state in flavour basis. flvf - The neutrino final flavour.
- Returns
- Neutrino oscillation probability
Definition at line 1114 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::P(), OscProb::PMNS_Base::Propagate(), and OscProb::PMNS_Base::SetPureState().
Referenced by AvgProb(), OscProb::PMNS_Base::AvgProb(), AvgProbLoE(), OscProb::PMNS_Base::AvgProbLoE(), and OscProb::PMNS_Base::Prob().
◆ Prob() [5/6]
|
virtualinherited |
Compute the probability of nu_in going to flvf for energy E
Compute the probability of nu_in going to flvf for a given energy in GeV.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
nu_in - The neutrino initial state in flavour basis. flvf - The neutrino final flavour. E - The neutrino energy in GeV
- Returns
- Neutrino oscillation probability
Definition at line 1138 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::Prob(), and OscProb::PMNS_Base::SetEnergy().
◆ Prob() [6/6]
|
virtualinherited |
Compute the probability of nu_in going to flvf for energy E and distance L
Compute the probability of nu_in going to flvf for a given energy in GeV and distance in km in a single path.
If the path sequence is not a single path, a new single path will be created and the previous sequence will be lost.
Don't use this if you want to propagate over multiple path segments.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
nu_in - The neutrino initial state in flavour basis. flvf - The neutrino final flavour. E - The neutrino energy in GeV L - The neutrino path length in km
- Returns
- Neutrino oscillation probability
Definition at line 1189 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::Prob(), OscProb::PMNS_Base::SetEnergy(), and OscProb::PMNS_Base::SetLength().
◆ ProbMatrix() [1/3]
|
virtualinherited |
Compute the probability matrix for the first nflvi and nflvf states.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
nflvi - The number of initial flavours in the matrix. nflvf - The number of final flavours in the matrix.
- Returns
- Neutrino oscillation probabilities
Reimplemented in OscProb::PMNS_DensityMatrix, OscProb::PMNS_DensityMatrix, OscProb::PMNS_NUNM, and OscProb::PMNS_OQS.
Definition at line 1387 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fNumNus, OscProb::PMNS_Base::fNuPaths, OscProb::PMNS_Base::fNuState, OscProb::PMNS_Base::PropagatePath(), and OscProb::PMNS_Base::ResetToFlavour().
Referenced by AvgProbMatrix(), OscProb::PMNS_Base::AvgProbMatrix(), AvgProbMatrixLoE(), OscProb::PMNS_Base::AvgProbMatrixLoE(), and OscProb::PMNS_Base::ProbMatrix().
◆ ProbMatrix() [2/3]
|
virtualinherited |
Compute the probability matrix for the first nflvi and nflvf states for a given energy in GeV.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
nflvi - The number of initial flavours in the matrix. nflvf - The number of final flavours in the matrix. E - The neutrino energy in GeV
- Returns
- Neutrino oscillation probabilities
Reimplemented in OscProb::PMNS_DensityMatrix.
Definition at line 1439 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::ProbMatrix(), and OscProb::PMNS_Base::SetEnergy().
◆ ProbMatrix() [3/3]
|
virtualinherited |
Compute the probability matrix for energy E and distance L
Compute the probability matrix for the first nflvi and nflvf states for a given energy in GeV and distance in km in a single path.
If the path sequence is not a single path, a new single path will be created and the previous sequence will be lost.
Don't use this if you want to propagate over multiple path segments.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
nflvi - The number of initial flavours in the matrix. nflvf - The number of final flavours in the matrix. E - The neutrino energy in GeV L - The neutrino path length in km
- Returns
- Neutrino oscillation probabilities
Reimplemented in OscProb::PMNS_DensityMatrix.
Definition at line 1468 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::ProbMatrix(), OscProb::PMNS_Base::SetEnergy(), and OscProb::PMNS_Base::SetLength().
◆ ProbVector() [1/6]
|
virtualinherited |
Compute the probabilities of flvi going to all flavours
Compute the probability of flvi going to all flavours.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
flvi - The neutrino starting flavour.
- Returns
- Neutrino oscillation probabilities
Definition at line 1272 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::GetProbVector(), OscProb::PMNS_Base::Propagate(), and OscProb::PMNS_Base::ResetToFlavour().
◆ ProbVector() [2/6]
|
virtualinherited |
Compute the probabilities of flvi going to all flavours for energy E
Compute the probability of flvi going to all flavours for a given energy in GeV.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
flvi - The neutrino starting flavour. E - The neutrino energy in GeV
- Returns
- Neutrino oscillation probability
Definition at line 1313 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::ProbVector(), and OscProb::PMNS_Base::SetEnergy().
◆ ProbVector() [3/6]
|
virtualinherited |
Compute the probabilities of flvi going to all flavours for energy E and distance L
Compute the probability of flvi going to all flavours for a given energy in GeV and distance in km in a single path.
If the path sequence is not a single path, a new single path will be created and the previous sequence will be lost.
Don't use this if you want to propagate over multiple path segments.
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
flvi - The neutrino starting flavour. E - The neutrino energy in GeV L - The neutrino path length in km
- Returns
- Neutrino oscillation probability
Definition at line 1365 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::ProbVector(), OscProb::PMNS_Base::SetEnergy(), and OscProb::PMNS_Base::SetLength().
◆ ProbVector() [4/6]
Compute the probabilities of nu_in going to all flavours
Compute the probability of nu_in going to all flavours.
- Parameters
-
nu_in - The neutrino initial state in flavour basis.
- Returns
- Neutrino oscillation probabilities
Definition at line 1250 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::GetProbVector(), OscProb::PMNS_Base::Propagate(), and OscProb::PMNS_Base::SetPureState().
Referenced by AvgProbVector(), OscProb::PMNS_Base::AvgProbVector(), AvgProbVectorLoE(), OscProb::PMNS_Base::AvgProbVectorLoE(), and OscProb::PMNS_Base::ProbVector().
◆ ProbVector() [5/6]
Compute the probabilities of nu_in going to all
Compute the probability of nu_in going to all flavours for a given energy in GeV.
- Parameters
-
nu_in - The neutrino initial state in flavour basis. E - The neutrino energy in GeV
- Returns
- Neutrino oscillation probabilities
Definition at line 1291 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::ProbVector(), and OscProb::PMNS_Base::SetEnergy().
◆ ProbVector() [6/6]
Compute the probabilities of nu_in going to all flavours for energy E and distance L
Compute the probability of nu_in going to all flavours for a given energy in GeV and distance in km in a single path.
If the path sequence is not a single path, a new single path will be created and the previous sequence will be lost.
Don't use this if you want to propagate over multiple path segments.
- Parameters
-
nu_in - The neutrino initial state in flavour basis. E - The neutrino energy in GeV L - The neutrino path length in km
- Returns
- Neutrino oscillation probabilities
Definition at line 1336 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::ProbVector(), OscProb::PMNS_Base::SetEnergy(), and OscProb::PMNS_Base::SetLength().
◆ Propagate()
|
protectedvirtualinherited |
Propagate neutrino state through full path
Reimplemented in OscProb::PMNS_NUNM, and OscProb::PMNS_OQS.
Definition at line 1018 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fNuPaths, and OscProb::PMNS_Base::PropagatePath().
Referenced by OscProb::PMNS_Base::Prob(), OscProb::PMNS_Base::ProbVector(), OscProb::PMNS_NUNM::Propagate(), and OscProb::PMNS_OQS::Propagate().
◆ PropagatePath()
|
protectedvirtualinherited |
Propagate the current neutrino state through a given path
- Parameters
-
p - A neutrino path segment
Reimplemented in OscProb::PMNS_Decay, OscProb::PMNS_Deco, OscProb::PMNS_Iter, OscProb::PMNS_NUNM, OscProb::PMNS_OQS, and OscProb::PMNS_DensityMatrix.
Definition at line 983 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fBuffer, OscProb::PMNS_Base::fEval, OscProb::PMNS_Base::fEvec, OscProb::PMNS_Base::fNumNus, OscProb::PMNS_Base::fNuState, OscProb::PMNS_Base::fPhases, OscProb::PMNS_Base::kKm2eV, OscProb::NuPath::length, OscProb::PMNS_Base::SetCurPath(), and OscProb::PMNS_Base::SolveHam().
Referenced by OscProb::PMNS_Base::ProbMatrix(), OscProb::PMNS_Base::Propagate(), OscProb::PMNS_Iter::PropagatePath(), and OscProb::PMNS_NUNM::PropagatePath().
◆ PropagatePathTaylor()
|
protectedvirtual |
Propagate the current neutrino state through a given path
- Parameters
-
p - A neutrino path segment
Definition at line 343 of file PMNS_Maltoni.cxx.
References BuildKcosT(), BuildKE(), fdcosT, fdInvE, OscProb::PMNS_Base::fEval, fKcosT, fKInvE, OscProb::PMNS_Base::fNumNus, OscProb::PMNS_Base::fPhases, OscProb::PMNS_Base::kKm2eV, OscProb::NuPath::length, MultiplicationRuleK(), MultiplicationRuleS(), rotateK(), rotateS(), OscProb::PMNS_Base::SetCurPath(), and OscProb::PMNS_Base::SolveHam().
Referenced by PropagateTaylor().
◆ PropagateTaylor()
|
protectedvirtual |
Propagate neutrino state through full path
Definition at line 330 of file PMNS_Maltoni.cxx.
References OscProb::PMNS_Base::fNuPaths, and PropagatePathTaylor().
Referenced by AvgAlgo(), AvgAlgoCosT(), AvgProb(), ExtrapolationProb(), ExtrapolationProbCosT(), and ExtrapolationProbLoE().
◆ ResetToFlavour()
|
protectedvirtualinherited |
Reset the neutrino state back to a pure flavour where it starts
Flavours are:
0 = nue, 1 = numu, 2 = nutau 3 = sterile_1, 4 = sterile_2, etc.
- Parameters
-
flv - The neutrino starting flavour.
Reimplemented in OscProb::PMNS_DensityMatrix.
Definition at line 1034 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fNumNus, OscProb::PMNS_Base::fNuState, OscProb::PMNS_Base::one, and OscProb::PMNS_Base::zero.
Referenced by OscProb::PMNS_Base::AvgProb(), OscProb::PMNS_Base::AvgProbLoE(), OscProb::PMNS_Base::AvgProbVector(), OscProb::PMNS_Base::AvgProbVectorLoE(), OscProb::PMNS_Base::GetMassEigenstate(), OscProb::PMNS_Base::PMNS_Base(), OscProb::PMNS_Base::Prob(), OscProb::PMNS_Base::ProbMatrix(), OscProb::PMNS_NUNM::ProbMatrix(), OscProb::PMNS_Base::ProbVector(), and OscProb::PMNS_DensityMatrix::ResetToFlavour().
◆ RotateDensityM()
|
protectedvirtual |
Apply rotation to the density matrix from or to the basis where V is diagonal
- Parameters
-
to_basis - Rotation from (false) or to (true) V - The matrix used for the denisty matrix rotation.
Definition at line 1173 of file PMNS_Maltoni.cxx.
References fdensityMatrix, and OscProb::PMNS_Base::fNumNus.
Referenced by AlgorithmDensityMatrix().
◆ RotateH()
|
protectedvirtualinherited |
Rotate the Hamiltonian by the angle theta_ij and phase delta_ij.
The rotations assume all off-diagonal elements with i > j are zero. This is correct if the order of rotations is chosen appropriately and it speeds up computation by skipping null terms
- Parameters
-
i,j - the indices of the rotation ij Ham - the Hamiltonian to be rotated
Definition at line 822 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fDelta, and OscProb::PMNS_Base::fTheta.
Referenced by OscProb::PMNS_Base::BuildHms(), OscProb::PMNS_Decay::BuildHms(), and OscProb::PMNS_SNSI::BuildHms().
◆ rotateK()
|
protectedvirtual |
Rotate the K matrix from mass to flavor basis
Definition at line 238 of file PMNS_Maltoni.cxx.
References OscProb::PMNS_Base::fEvec, fKflavor, fKmass, and OscProb::PMNS_Base::fNumNus.
Referenced by PropagatePathTaylor().
◆ rotateS()
|
protectedvirtual |
Rotate the S matrix for the current layer from mass to flavor basis
Definition at line 216 of file PMNS_Maltoni.cxx.
References OscProb::PMNS_Base::fEvec, OscProb::PMNS_Base::fNumNus, OscProb::PMNS_Base::fPhases, and fSflavor.
Referenced by PropagatePathTaylor().
◆ RotateState()
|
protectedvirtualinherited |
Rotate the neutrino state by the angle theta_ij and phase delta_ij.
- Parameters
-
i,j - the indices of the rotation ij
Definition at line 760 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fDelta, OscProb::PMNS_Base::fNuState, and OscProb::PMNS_Base::fTheta.
Referenced by OscProb::PMNS_Base::GetMassEigenstate().
◆ SetAngle()
|
virtualinherited |
Set the mixing angle theta_ij in radians.
Requires that i<j. Will notify you if input is wrong. If i>j, will assume reverse order and swap i and j.
This will check if value is changing to keep track of whether the hamiltonian needs to be rebuilt.
- Parameters
-
i,j - the indices of theta_ij th - the value of theta_ij
Definition at line 539 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fBuiltHms, OscProb::PMNS_Base::fNumNus, and OscProb::PMNS_Base::fTheta.
Referenced by GetSterile(), OscProb::PMNS_Decay::SetMix(), OscProb::PMNS_Fast::SetMix(), SetNominalPars(), and OscProb::PMNS_Base::SetStdPars().
◆ SetAtt() [1/2]
|
protectedvirtualinherited |
Set some single path attribute.
An auxiliary function to set individual attributes in a single path.
If the path sequence is not a single path, a new single path will be created and the previous sequence will be lost. Use with care.
- Parameters
-
att - The value of the attribute idx - The index of the attribute (0,1,2,3) = (L, Rho, Z/A, Layer)
Definition at line 364 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fNuPaths, and OscProb::PMNS_Base::SetStdPath().
Referenced by OscProb::PMNS_Base::SetDensity(), OscProb::PMNS_Base::SetLayers(), OscProb::PMNS_Base::SetLength(), and OscProb::PMNS_Base::SetZoA().
◆ SetAtt() [2/2]
|
protectedvirtualinherited |
Set all values of a path attribute.
An auxiliary function to set individual attributes in a path sequence.
If the path sequence is of a different size, a new path sequence will be created and the previous sequence will be lost. Use with care.
- Parameters
-
att - The values of the attribute idx - The index of the attribute (0,1,2,3) = (L, Rho, Z/A, Layer)
Definition at line 427 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::AddPath(), OscProb::PMNS_Base::ClearPath(), OscProb::NuPath::density, OscProb::PMNS_Base::fNuPaths, OscProb::NuPath::layer, OscProb::NuPath::length, OscProb::PMNS_Base::SetStdPath(), and OscProb::NuPath::zoa.
◆ SetAvgProbPrec()
|
virtualinherited |
Set the precision for the AvgProb method
- Parameters
-
prec - AvgProb precision
Definition at line 1962 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fAvgProbPrec.
Referenced by OscProb::PMNS_Base::PMNS_Base(), and PMNS_Maltoni().
◆ SetCosT()
|
protectedvirtual |
Set neutrino zenith angle.
- Parameters
-
cosT - The cosine of the zenith angle
Definition at line 92 of file PMNS_Maltoni.cxx.
References fcosT.
Referenced by AvgAlgo(), AvgAlgoCosT(), AvgProb(), and ExtrapolationProbCosT().
◆ SetCurPath()
|
protectedvirtualinherited |
Set the path currentlyin use by the class.
This will be used to know what path to propagate through next.
It will also check if values are changing to keep track of whether the eigensystem needs to be recomputed.
- Parameters
-
p - A neutrino path segment
Definition at line 274 of file PMNS_Base.cxx.
References OscProb::NuPath::density, OscProb::PMNS_Base::fGotES, OscProb::PMNS_Base::fPath, and OscProb::NuPath::zoa.
Referenced by OscProb::PMNS_Base::AvgProbLoE(), OscProb::PMNS_Base::AvgProbMatrixLoE(), OscProb::PMNS_Base::AvgProbVectorLoE(), OscProb::PMNS_OQS::BuildHVMB(), OscProb::PMNS_Base::ConvertEtoLoE(), ExtrapolationProbLoE(), OscProb::PMNS_Base::PropagatePath(), OscProb::PMNS_Decay::PropagatePath(), OscProb::PMNS_Deco::PropagatePath(), and PropagatePathTaylor().
◆ SetDelta()
|
virtualinherited |
Set the CP phase delta_ij in radians.
Requires that i+1<j. Will notify you if input is wrong. If i>j, will assume reverse order and swap i and j.
This will check if value is changing to keep track of whether the hamiltonian needs to be rebuilt.
- Parameters
-
i,j - the indices of delta_ij delta - the value of delta_ij
Definition at line 602 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fBuiltHms, OscProb::PMNS_Base::fDelta, and OscProb::PMNS_Base::fNumNus.
Referenced by OscProb::PMNS_Decay::SetMix(), OscProb::PMNS_Fast::SetMix(), and SetNominalPars().
◆ SetDensity() [1/2]
|
virtualinherited |
Set single path density.
If the path sequence is not a single path, a new single path will be created and the previous sequence will be lost. Use with care.
- Parameters
-
rho - The density of the path segment in g/cm^3
Definition at line 402 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::SetAtt().
◆ SetDensity() [2/2]
|
virtualinherited |
Set multiple path densities.
If the path sequence is of a different size, a new path sequence will be created and the previous sequence will be lost. Use with care.
- Parameters
-
rho - The densities of the path segments in g/cm^3
Definition at line 497 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::SetAtt().
◆ SetDm()
|
virtualinherited |
Set the mass-splitting dm_j1 = (m_j^2 - m_1^2) in eV^2
Requires that j>1. Will notify you if input is wrong.
This will check if value is changing to keep track of whether the hamiltonian needs to be rebuilt.
- Parameters
-
j - the index of dm_j1 dm - the value of dm_j1
Definition at line 674 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fBuiltHms, OscProb::PMNS_Base::fDm, and OscProb::PMNS_Base::fNumNus.
Referenced by GetSterile(), OscProb::PMNS_Decay::SetDeltaMsqrs(), OscProb::PMNS_Fast::SetDeltaMsqrs(), SetNominalPars(), and OscProb::PMNS_Base::SetStdPars().
◆ SetEnergy()
|
virtualinherited |
Set neutrino energy in GeV.
This will check if value is changing to keep track of whether the eigensystem needs to be recomputed.
- Parameters
-
E - The neutrino energy in GeV
Definition at line 226 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fEnergy, and OscProb::PMNS_Base::fGotES.
Referenced by AvgAlgo(), AvgAlgoCosT(), AvgProb(), OscProb::PMNS_Base::AvgProbLoE(), OscProb::PMNS_Base::AvgProbMatrixLoE(), OscProb::PMNS_Base::AvgProbVectorLoE(), ExtrapolationProb(), ExtrapolationProbLoE(), OscProb::PMNS_Base::PMNS_Base(), OscProb::PMNS_Base::Prob(), OscProb::PMNS_Base::ProbMatrix(), and OscProb::PMNS_Base::ProbVector().
◆ SetIsNuBar()
|
virtualinherited |
Set anti-neutrino flag.
This will check if value is changing to keep track of whether the eigensystem needs to be recomputed.
- Parameters
-
isNuBar - Set to true for anti-neutrino and false for neutrino.
Reimplemented in OscProb::PMNS_Decay, OscProb::PMNS_Iter, and OscProb::PMNS_OQS.
Definition at line 243 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fGotES, and OscProb::PMNS_Base::fIsNuBar.
Referenced by CheckProb(), OscProb::PMNS_Base::PMNS_Base(), SaveTestFile(), and OscProb::PMNS_OQS::SetIsNuBar().
◆ SetIsOscProbAvg()
|
virtual |
Set method used for averaging.
- Parameters
-
isOscProbAvg - OscProb (true) or Maltoni (false)
Reimplemented in OscProb::PMNS_DensityMatrix, OscProb::PMNS_LIV, OscProb::PMNS_NUNM, and OscProb::PMNS_SNSI.
Definition at line 70 of file PMNS_Maltoni.cxx.
References fIsOscProbAvg.
Referenced by PMNS_Maltoni().
◆ SetLayers()
|
virtualinherited |
Set multiple path layer indices.
If the path sequence is of a different size, a new path sequence will be created and the previous sequence will be lost. Use with care.
- Parameters
-
lay - Indices to identify the layer types (e.g. earth inner core)
Definition at line 519 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::SetAtt().
◆ SetLength() [1/2]
|
virtualinherited |
Set the length for a single path.
If the path sequence is not a single path, a new single path will be created and the previous sequence will be lost. Use with care.
- Parameters
-
L - The length of the path segment in km
Definition at line 391 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::SetAtt().
Referenced by OscProb::PMNS_Base::Prob(), OscProb::PMNS_Base::ProbMatrix(), and OscProb::PMNS_Base::ProbVector().
◆ SetLength() [2/2]
|
virtualinherited |
Set multiple path lengths.
If the path sequence is of a different size, a new path sequence will be created and the previous sequence will be lost. Use with care.
- Parameters
-
L - The lengths of the path segments in km
Definition at line 486 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::SetAtt().
◆ SetMaxCache()
|
virtualinherited |
Set maximum number of cached eigensystems. Finding eigensystems can become slow and take up memory. This protects the cache from becoming too large.
- Parameters
-
mc - Max cache size (default: 1e6)
Definition at line 128 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fMaxCache.
◆ SetPath() [1/3]
|
virtualinherited |
Set a single path defining attributes directly.
This destroys the current path sequence and creates a new first path.
- Parameters
-
length - The length of the path segment in km density - The density of the path segment in g/cm^3 zoa - The effective Z/A of the path segment layer - An index to identify the layer type (e.g. earth inner core)
Definition at line 347 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::SetPath().
◆ SetPath() [2/3]
|
virtualinherited |
Set a single path.
This destroys the current path sequence and creates a new first path.
- Parameters
-
p - A neutrino path segment
Definition at line 330 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::AddPath(), and OscProb::PMNS_Base::ClearPath().
Referenced by AvgAlgo(), AvgAlgoCosT(), AvgProb(), ExtrapolationProbCosT(), OscProb::PMNS_Base::SetPath(), OscProb::PMNS_Base::SetStdPath(), and SetTestPath().
◆ SetPath() [3/3]
|
virtualinherited |
Set vector of neutrino paths.
- Parameters
-
paths - A sequence of neutrino paths
Definition at line 294 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fNuPaths.
◆ SetPremModel()
|
virtual |
Set the earth model to be used.
This is done to get access to the PremLayer list to be used in the LnDerivative() function.
- Parameters
-
prem - The earth model used
Definition at line 84 of file PMNS_Maltoni.cxx.
References fPrem.
◆ SetPureState()
|
protectedvirtualinherited |
Set the initial state from a pure state
- Parameters
-
nu_in - The neutrino initial state in flavour basis.
Reimplemented in OscProb::PMNS_DensityMatrix.
Definition at line 1070 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fNumNus, and OscProb::PMNS_Base::fNuState.
Referenced by OscProb::PMNS_Base::Prob(), and OscProb::PMNS_Base::ProbVector().
◆ SetStdPars()
|
virtualinherited |
Set standard oscillation parameters from PDG 2015.
For two neutrinos, Dm is set to the muon disappearance effective mass-splitting and mixing angle.
Definition at line 177 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fNumNus, OscProb::PMNS_Base::SetAngle(), and OscProb::PMNS_Base::SetDm().
Referenced by OscProb::PMNS_Base::PMNS_Base().
◆ SetStdPath()
|
virtualinherited |
Set standard single path.
Length is 1000 km, so ~2 GeV peak energy.
Density is approximate from CRUST2.0 (~2.8 g/cm^3). Z/A is set to a round 0.5.
Definition at line 205 of file PMNS_Base.cxx.
References OscProb::NuPath::density, OscProb::NuPath::layer, OscProb::NuPath::length, OscProb::PMNS_Base::SetPath(), and OscProb::NuPath::zoa.
Referenced by OscProb::PMNS_Base::PMNS_Base(), OscProb::PMNS_LIV::PMNS_LIV(), OscProb::PMNS_NSI::PMNS_NSI(), OscProb::PMNS_NUNM::PMNS_NUNM(), and OscProb::PMNS_Base::SetAtt().
◆ SetUseCache()
|
virtualinherited |
Turn on/off caching of eigensystems. This can save a lot of CPU time by avoiding recomputing eigensystems if we've already seen them recently. Especially useful when running over multiple earth layers and even more if multiple baselines will be computed, e.g. for atmospheric neutrinos.
- Parameters
-
u - flag to set caching on (default: true)
Definition at line 105 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fUseCache.
Referenced by OscProb::PMNS_Base::PMNS_Base().
◆ SetwidthBin()
|
protectedvirtual |
Set bin widths.
- Parameters
-
dE - The width of the energy bin in GeV dcosT - The width of the cos(theta_z) bin
Definition at line 101 of file PMNS_Maltoni.cxx.
References fdcosT, and fdInvE.
Referenced by AvgAlgo(), AvgAlgoCosT(), AvgProb(), ExtrapolationProb(), ExtrapolationProbCosT(), ExtrapolationProbLoE(), and PMNS_Maltoni().
◆ SetZoA() [1/2]
|
virtualinherited |
Set single path Z/A.
If the path sequence is not a single path, a new single path will be created and the previous sequence will be lost. Use with care.
- Parameters
-
zoa - The effective Z/A of the path segment
Definition at line 413 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::SetAtt().
◆ SetZoA() [2/2]
|
virtualinherited |
Set multiple path Z/A values.
If the path sequence is of a different size, a new path sequence will be created and the previous sequence will be lost. Use with care.
- Parameters
-
zoa - The effective Z/A of the path segments
Definition at line 508 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::SetAtt().
◆ SolveHam()
|
protectedpure virtualinherited |
Solve the full Hamiltonian
Not implemented in base class. Solve the full Hamiltonian for eigenvectors and eigenvalues
Implemented in OscProb::PMNS_Decay, OscProb::PMNS_Fast, OscProb::PMNS_Iter, OscProb::PMNS_LIV, and OscProb::PMNS_Sterile.
Referenced by OscProb::PMNS_Base::GetDmEff(), OscProb::PMNS_Base::GetSamplePoints(), OscProb::PMNS_Base::PropagatePath(), and PropagatePathTaylor().
◆ SolveK()
Solve one K matrix for eigenvectors and eigenvalues.
This is using the Eigen methods appropriate for each matrix size.
- Parameters
-
K - The K matrix lambda - The eigenvalues of K V - The eigenvectors of K
Definition at line 395 of file PMNS_Maltoni.cxx.
References OscProb::PMNS_Base::fNumNus.
Referenced by AvgAlgo(), AvgAlgoCosT(), AvgProb(), ExtrapolationProb(), ExtrapolationProbCosT(), and ExtrapolationProbLoE().
◆ TemplateSolver()
|
protected |
Auxiliary function to convert a generic Xcd matrix into a specific size so Eigen can optimize the eigensolver method used.
- Parameters
-
K - The K matrix lambda - The eigenvalues of K V - The eigenvectors of K
Definition at line 415 of file PMNS_Maltoni.cxx.
References OscProb::PMNS_Base::fNumNus.
◆ TryCache()
|
protectedvirtualinherited |
Try to find a cached version of this eigensystem.
Definition at line 134 of file PMNS_Base.cxx.
References OscProb::PMNS_Base::fEnergy, OscProb::PMNS_Base::fEval, OscProb::PMNS_Base::fEvec, OscProb::PMNS_Base::fIsNuBar, OscProb::PMNS_Base::fMixCache, OscProb::PMNS_Base::fNumNus, OscProb::PMNS_Base::fPath, OscProb::PMNS_Base::fProbe, OscProb::PMNS_Base::fUseCache, and OscProb::EigenPoint::SetVars().
Referenced by OscProb::PMNS_Sterile::SolveHam(), and OscProb::PMNS_Fast::SolveHamMatter().
◆ UpdateHam()
|
protectedpure virtual |
Implemented in OscProb::PMNS_Fast, OscProb::PMNS_LIV, OscProb::PMNS_NSI, OscProb::PMNS_NUNM, OscProb::PMNS_SNSI, and OscProb::PMNS_Sterile.
Referenced by BuildKcosT().
Member Data Documentation
◆ fAvgProbPrec
|
protectedinherited |
Definition at line 305 of file PMNS_Base.h.
Referenced by OscProb::PMNS_Base::GetSamplePoints(), GetSamplePointsAvgClass(), and OscProb::PMNS_Base::SetAvgProbPrec().
◆ fBuffer
|
protectedinherited |
Definition at line 287 of file PMNS_Base.h.
Referenced by OscProb::PMNS_Base::PropagatePath(), and OscProb::PMNS_Decay::PropagatePath().
◆ fBuiltHms
|
protectedinherited |
Definition at line 298 of file PMNS_Base.h.
Referenced by OscProb::PMNS_Base::BuildHms(), OscProb::PMNS_Decay::BuildHms(), OscProb::PMNS_OQS::BuildHms(), OscProb::PMNS_SNSI::BuildHms(), OscProb::PMNS_Decay::SetAlpha2(), OscProb::PMNS_Decay::SetAlpha3(), OscProb::PMNS_Base::SetAngle(), OscProb::PMNS_Base::SetDelta(), OscProb::PMNS_Base::SetDm(), OscProb::PMNS_Decay::SetIsNuBar(), OscProb::PMNS_Iter::SetIsNuBar(), OscProb::PMNS_OQS::SetIsNuBar(), OscProb::PMNS_SNSI::SetLowestMass(), and OscProb::PMNS_Iter::SolveHam().
◆ fCachePrec
|
protectedinherited |
Definition at line 302 of file PMNS_Base.h.
◆ fcosT
|
protected |
Definition at line 184 of file PMNS_Maltoni.h.
Referenced by LnDerivative(), and SetCosT().
◆ fdcosT
|
protected |
Definition at line 187 of file PMNS_Maltoni.h.
Referenced by AlgorithmDensityMatrix(), AvgAlgoCosT(), AvgProb(), ExtrapolationProbCosT(), PropagatePathTaylor(), and SetwidthBin().
◆ fDelta
|
protectedinherited |
Definition at line 281 of file PMNS_Base.h.
Referenced by OscProb::PMNS_Base::GetDelta(), OscProb::PMNS_Base::RotateH(), OscProb::PMNS_Base::RotateState(), OscProb::PMNS_Base::SetDelta(), and OscProb::PMNS_Fast::SetVacuumEigensystem().
◆ fdensityMatrix
|
protected |
Definition at line 206 of file PMNS_Maltoni.h.
Referenced by AlgorithmDensityMatrix(), HadamardProduct(), InitializeTaylorsVectors(), and RotateDensityM().
◆ fDetRadius
|
protected |
Definition at line 211 of file PMNS_Maltoni.h.
Referenced by AvgAlgo(), AvgAlgoCosT(), AvgProb(), ExtrapolationProbCosT(), InitializeTaylorsVectors(), and LnDerivative().
◆ fdInvE
|
protected |
Definition at line 186 of file PMNS_Maltoni.h.
Referenced by AlgorithmDensityMatrix(), AvgProbMatrixLoE(), AvgProbVectorLoE(), ExtrapolationProb(), PropagatePathTaylor(), and SetwidthBin().
◆ fdl
|
protected |
Definition at line 210 of file PMNS_Maltoni.h.
Referenced by InitializeTaylorsVectors(), and LnDerivative().
◆ fDm
|
protectedinherited |
Definition at line 279 of file PMNS_Base.h.
Referenced by OscProb::PMNS_Base::BuildHms(), OscProb::PMNS_Decay::BuildHms(), OscProb::PMNS_SNSI::BuildHms(), OscProb::PMNS_OQS::BuildHVMB(), OscProb::PMNS_Base::GetDm(), OscProb::PMNS_Base::GetDmEff(), OscProb::PMNS_DensityMatrix::ProbMatrix(), OscProb::PMNS_Deco::PropagatePath(), OscProb::PMNS_Iter::PropagatePath(), OscProb::PMNS_Base::SetDm(), and OscProb::PMNS_Fast::SetVacuumEigensystem().
◆ fEnergy
|
protectedinherited |
Definition at line 292 of file PMNS_Base.h.
Referenced by OscProb::PMNS_OQS::BuildHVMB(), OscProb::PMNS_OQS::BuildM(), OscProb::PMNS_Base::FillCache(), OscProb::PMNS_Base::GetDmEff(), OscProb::PMNS_Base::GetEnergy(), OscProb::PMNS_Base::GetSamplePoints(), OscProb::PMNS_Deco::PropagatePath(), OscProb::PMNS_Iter::PropagatePath(), OscProb::PMNS_Base::SetEnergy(), OscProb::PMNS_Fast::SetVacuumEigensystem(), OscProb::PMNS_Iter::SolveHam(), OscProb::PMNS_Base::TryCache(), OscProb::PMNS_Decay::UpdateHam(), OscProb::PMNS_Fast::UpdateHam(), OscProb::PMNS_LIV::UpdateHam(), OscProb::PMNS_NSI::UpdateHam(), OscProb::PMNS_NUNM::UpdateHam(), OscProb::PMNS_SNSI::UpdateHam(), and OscProb::PMNS_Sterile::UpdateHam().
◆ fEval
|
protectedinherited |
Definition at line 289 of file PMNS_Base.h.
Referenced by BuildKE(), OscProb::PMNS_Base::FillCache(), OscProb::PMNS_Base::GetDmEff(), OscProb::PMNS_Base::GetSamplePoints(), OscProb::PMNS_Base::PropagatePath(), OscProb::PMNS_Deco::PropagatePath(), PropagatePathTaylor(), OscProb::PMNS_Fast::SetVacuumEigensystem(), OscProb::PMNS_Sterile::SolveEigenSystem(), OscProb::PMNS_Decay::SolveHam(), OscProb::PMNS_Iter::SolveHam(), OscProb::PMNS_Fast::SolveHamMatter(), and OscProb::PMNS_Base::TryCache().
◆ fEvec
|
protectedinherited |
Definition at line 290 of file PMNS_Base.h.
Referenced by OscProb::PMNS_OQS::BuildHms(), BuildKE(), OscProb::PMNS_Base::FillCache(), OscProb::PMNS_Base::PropagatePath(), OscProb::PMNS_Deco::PropagatePath(), rotateK(), rotateS(), OscProb::PMNS_Fast::SetVacuumEigensystem(), OscProb::PMNS_Sterile::SolveEigenSystem(), OscProb::PMNS_Fast::SolveHamMatter(), and OscProb::PMNS_Base::TryCache().
◆ fevolutionMatrixS
|
protected |
Evolution matrix S for reference energy and angle for the entire path
Definition at line 200 of file PMNS_Maltoni.h.
Referenced by AlgorithmDensityMatrix(), AvgFormula(), AvgFormulaExtrapolation(), InitializeTaylorsVectors(), MultiplicationRuleK(), and MultiplicationRuleS().
◆ fGotES
|
protectedinherited |
Definition at line 299 of file PMNS_Base.h.
Referenced by OscProb::PMNS_Base::BuildHms(), OscProb::PMNS_Decay::BuildHms(), OscProb::PMNS_SNSI::BuildHms(), OscProb::PMNS_NUNM::SetAlpha(), OscProb::PMNS_LIV::SetaT(), OscProb::PMNS_NSI::SetCoupByIndex(), OscProb::PMNS_LIV::SetcT(), OscProb::PMNS_Base::SetCurPath(), OscProb::PMNS_Base::SetEnergy(), OscProb::PMNS_NSI::SetEps(), OscProb::PMNS_NUNM::SetFracVnc(), OscProb::PMNS_Base::SetIsNuBar(), OscProb::PMNS_Decay::SetIsNuBar(), OscProb::PMNS_Decay::SolveHam(), OscProb::PMNS_Iter::SolveHam(), OscProb::PMNS_Sterile::SolveHam(), and OscProb::PMNS_Fast::SolveHamMatter().
◆ fHam
|
protected |
Definition at line 217 of file PMNS_Maltoni.h.
Referenced by BuildKcosT().
◆ fHms
|
protectedinherited |
Definition at line 284 of file PMNS_Base.h.
Referenced by OscProb::PMNS_Base::BuildHms(), OscProb::PMNS_Decay::BuildHms(), OscProb::PMNS_SNSI::BuildHms(), BuildKE(), OscProb::PMNS_Decay::UpdateHam(), OscProb::PMNS_Fast::UpdateHam(), OscProb::PMNS_LIV::UpdateHam(), OscProb::PMNS_NSI::UpdateHam(), OscProb::PMNS_NUNM::UpdateHam(), OscProb::PMNS_SNSI::UpdateHam(), and OscProb::PMNS_Sterile::UpdateHam().
◆ fIsNuBar
|
protectedinherited |
Definition at line 293 of file PMNS_Base.h.
Referenced by OscProb::PMNS_Decay::BuildHms(), OscProb::PMNS_OQS::BuildHVMB(), BuildKE(), OscProb::PMNS_Base::FillCache(), OscProb::PMNS_Base::GetIsNuBar(), OscProb::PMNS_Iter::SetExpVL(), OscProb::PMNS_Base::SetIsNuBar(), OscProb::PMNS_Decay::SetIsNuBar(), OscProb::PMNS_Iter::SetIsNuBar(), OscProb::PMNS_OQS::SetIsNuBar(), OscProb::PMNS_Fast::SetVacuumEigensystem(), OscProb::PMNS_Base::TryCache(), OscProb::PMNS_Decay::UpdateHam(), OscProb::PMNS_Fast::UpdateHam(), OscProb::PMNS_LIV::UpdateHam(), OscProb::PMNS_NSI::UpdateHam(), OscProb::PMNS_NUNM::UpdateHam(), OscProb::PMNS_SNSI::UpdateHam(), and OscProb::PMNS_Sterile::UpdateHam().
◆ fIsOscProbAvg
|
protected |
Definition at line 219 of file PMNS_Maltoni.h.
Referenced by AvgProb(), AvgProbLoE(), AvgProbMatrix(), AvgProbMatrixLoE(), AvgProbVector(), AvgProbVectorLoE(), OscProb::PMNS_DensityMatrix::SetIsOscProbAvg(), OscProb::PMNS_LIV::SetIsOscProbAvg(), SetIsOscProbAvg(), OscProb::PMNS_NUNM::SetIsOscProbAvg(), and OscProb::PMNS_SNSI::SetIsOscProbAvg().
◆ fKcosT
|
protected |
Definition at line 196 of file PMNS_Maltoni.h.
Referenced by AvgAlgo(), AvgAlgoCosT(), AvgProb(), ExtrapolationProbCosT(), InitializeTaylorsVectors(), and PropagatePathTaylor().
◆ fKflavor
|
protected |
Definition at line 204 of file PMNS_Maltoni.h.
Referenced by BuildKcosT(), InitializeTaylorsVectors(), MultiplicationRuleK(), and rotateK().
◆ fKInvE
|
protected |
Definition at line 194 of file PMNS_Maltoni.h.
Referenced by AvgAlgo(), ExtrapolationProb(), ExtrapolationProbLoE(), InitializeTaylorsVectors(), and PropagatePathTaylor().
◆ fKmass
|
protected |
Definition at line 203 of file PMNS_Maltoni.h.
Referenced by BuildKE(), InitializeTaylorsVectors(), and rotateK().
◆ flambdaCosT
|
protected |
Definition at line 190 of file PMNS_Maltoni.h.
Referenced by AlgorithmDensityMatrix(), AvgAlgo(), AvgAlgoCosT(), AvgProb(), ExtrapolationProbCosT(), and InitializeTaylorsVectors().
◆ flambdaInvE
|
protected |
Definition at line 189 of file PMNS_Maltoni.h.
Referenced by AlgorithmDensityMatrix(), AvgAlgo(), AvgProbMatrixLoE(), AvgProbVectorLoE(), ExtrapolationProb(), ExtrapolationProbLoE(), and InitializeTaylorsVectors().
◆ fLayer
|
protected |
Definition at line 209 of file PMNS_Maltoni.h.
Referenced by InitializeTaylorsVectors(), and LnDerivative().
◆ fMaxCache
|
protectedinherited |
Definition at line 303 of file PMNS_Base.h.
Referenced by OscProb::PMNS_Base::FillCache(), and OscProb::PMNS_Base::SetMaxCache().
◆ fminRsq
|
protected |
Definition at line 212 of file PMNS_Maltoni.h.
Referenced by AvgAlgo(), AvgAlgoCosT(), AvgProb(), ExtrapolationProbCosT(), and LnDerivative().
◆ fMixCache
|
protectedinherited |
Definition at line 307 of file PMNS_Base.h.
Referenced by OscProb::PMNS_Base::ClearCache(), OscProb::PMNS_Base::FillCache(), and OscProb::PMNS_Base::TryCache().
◆ fNumNus
|
protectedinherited |
Definition at line 277 of file PMNS_Base.h.
Referenced by OscProb::PMNS_NUNM::ApplyAlphaDagger(), AvgFormula(), AvgFormulaExtrapolation(), AvgProbVector(), OscProb::PMNS_Base::AvgProbVector(), AvgProbVectorLoE(), OscProb::PMNS_Base::AvgProbVectorLoE(), OscProb::PMNS_Base::BuildHms(), OscProb::PMNS_Decay::BuildHms(), OscProb::PMNS_SNSI::BuildHms(), BuildKcosT(), BuildKE(), OscProb::PMNS_Base::FillCache(), OscProb::PMNS_NUNM::GetAlpha(), OscProb::PMNS_Base::GetAngle(), OscProb::PMNS_LIV::GetaT(), OscProb::PMNS_LIV::GetcT(), OscProb::PMNS_Base::GetDelta(), OscProb::PMNS_Base::GetDm(), OscProb::PMNS_Base::GetDmEff(), OscProb::PMNS_NSI::GetEps(), OscProb::PMNS_Base::GetMassEigenstate(), OscProb::PMNS_Base::GetProbVector(), OscProb::PMNS_Base::GetSamplePoints(), HadamardProduct(), InitializeTaylorsVectors(), MultiplicationRuleK(), MultiplicationRuleS(), OscProb::PMNS_Base::P(), OscProb::PMNS_DensityMatrix::P(), OscProb::PMNS_Base::PMNS_Base(), OscProb::PMNS_Decay::PMNS_Decay(), OscProb::PMNS_Base::ProbMatrix(), OscProb::PMNS_NUNM::ProbMatrix(), OscProb::PMNS_OQS::ProbMatrix(), OscProb::PMNS_DensityMatrix::ProbMatrix(), OscProb::PMNS_Base::PropagatePath(), OscProb::PMNS_Decay::PropagatePath(), OscProb::PMNS_Deco::PropagatePath(), OscProb::PMNS_Iter::PropagatePath(), PropagatePathTaylor(), OscProb::PMNS_Base::ResetToFlavour(), OscProb::PMNS_DensityMatrix::ResetToFlavour(), RotateDensityM(), rotateK(), rotateS(), OscProb::PMNS_DensityMatrix::RotateState(), OscProb::PMNS_NUNM::SetAlpha(), OscProb::PMNS_Base::SetAngle(), OscProb::PMNS_LIV::SetaT(), OscProb::PMNS_LIV::SetcT(), OscProb::PMNS_Base::SetDelta(), OscProb::PMNS_Base::SetDm(), OscProb::PMNS_NSI::SetEps(), OscProb::PMNS_DensityMatrix::SetInitialRho(), OscProb::PMNS_Base::SetPureState(), OscProb::PMNS_DensityMatrix::SetPureState(), OscProb::PMNS_Base::SetStdPars(), OscProb::PMNS_Sterile::SolveEigenSystem(), OscProb::PMNS_Iter::SolveHam(), OscProb::PMNS_Sterile::SolveHam(), OscProb::PMNS_Fast::SolveHamMatter(), SolveK(), TemplateSolver(), OscProb::PMNS_Base::TryCache(), OscProb::PMNS_Decay::UpdateHam(), OscProb::PMNS_Fast::UpdateHam(), OscProb::PMNS_LIV::UpdateHam(), OscProb::PMNS_NSI::UpdateHam(), OscProb::PMNS_NUNM::UpdateHam(), OscProb::PMNS_SNSI::UpdateHam(), and OscProb::PMNS_Sterile::UpdateHam().
◆ fNuPaths
|
protectedinherited |
Definition at line 295 of file PMNS_Base.h.
Referenced by OscProb::PMNS_Base::AddPath(), AvgProb(), OscProb::PMNS_Base::AvgProb(), AvgProbLoE(), OscProb::PMNS_Base::AvgProbLoE(), AvgProbMatrix(), OscProb::PMNS_Base::AvgProbMatrix(), AvgProbMatrixLoE(), OscProb::PMNS_Base::AvgProbMatrixLoE(), AvgProbVector(), OscProb::PMNS_Base::AvgProbVector(), AvgProbVectorLoE(), OscProb::PMNS_Base::AvgProbVectorLoE(), OscProb::PMNS_Base::ClearPath(), OscProb::PMNS_Base::ConvertEtoLoE(), ExtrapolationProbLoE(), OscProb::PMNS_Base::GetPath(), OscProb::PMNS_Base::ProbMatrix(), OscProb::PMNS_DensityMatrix::ProbMatrix(), OscProb::PMNS_NUNM::ProbMatrix(), OscProb::PMNS_OQS::ProbMatrix(), OscProb::PMNS_Base::Propagate(), PropagateTaylor(), OscProb::PMNS_Base::SetAtt(), and OscProb::PMNS_Base::SetPath().
◆ fNuState
|
protectedinherited |
Definition at line 283 of file PMNS_Base.h.
Referenced by OscProb::PMNS_Base::AvgProb(), OscProb::PMNS_Base::AvgProbLoE(), OscProb::PMNS_Base::AvgProbVector(), OscProb::PMNS_Base::AvgProbVectorLoE(), OscProb::PMNS_Base::GetMassEigenstate(), OscProb::PMNS_Base::P(), OscProb::PMNS_Base::ProbMatrix(), OscProb::PMNS_NUNM::ProbMatrix(), OscProb::PMNS_NUNM::Propagate(), OscProb::PMNS_Base::PropagatePath(), OscProb::PMNS_Decay::PropagatePath(), OscProb::PMNS_Iter::PropMatter(), OscProb::PMNS_Base::ResetToFlavour(), OscProb::PMNS_Base::RotateState(), and OscProb::PMNS_Base::SetPureState().
◆ fPath
|
protectedinherited |
Definition at line 296 of file PMNS_Base.h.
Referenced by AvgProbLoE(), OscProb::PMNS_Base::AvgProbLoE(), AvgProbMatrixLoE(), OscProb::PMNS_Base::AvgProbMatrixLoE(), AvgProbVectorLoE(), OscProb::PMNS_Base::AvgProbVectorLoE(), OscProb::PMNS_OQS::BuildHVMB(), OscProb::PMNS_Base::ConvertEtoLoE(), ExtrapolationProbLoE(), OscProb::PMNS_Base::FillCache(), OscProb::PMNS_NSI::GetZoACoup(), OscProb::PMNS_Base::SetCurPath(), OscProb::PMNS_Fast::SolveHam(), OscProb::PMNS_Base::TryCache(), OscProb::PMNS_Decay::UpdateHam(), OscProb::PMNS_Fast::UpdateHam(), OscProb::PMNS_LIV::UpdateHam(), OscProb::PMNS_NSI::UpdateHam(), OscProb::PMNS_NUNM::UpdateHam(), OscProb::PMNS_SNSI::UpdateHam(), and OscProb::PMNS_Sterile::UpdateHam().
◆ fPhases
|
protectedinherited |
Definition at line 286 of file PMNS_Base.h.
Referenced by OscProb::PMNS_Base::PropagatePath(), PropagatePathTaylor(), and rotateS().
◆ fPrem
|
protected |
Definition at line 215 of file PMNS_Maltoni.h.
Referenced by AvgAlgo(), AvgAlgoCosT(), AvgProb(), ExtrapolationProbCosT(), InitializeTaylorsVectors(), LnDerivative(), PMNS_Maltoni(), and SetPremModel().
◆ fProbe
|
protectedinherited |
Definition at line 308 of file PMNS_Base.h.
Referenced by OscProb::PMNS_Base::FillCache(), and OscProb::PMNS_Base::TryCache().
◆ fSflavor
|
protected |
Definition at line 202 of file PMNS_Maltoni.h.
Referenced by InitializeTaylorsVectors(), MultiplicationRuleS(), and rotateS().
◆ fTheta
|
protectedinherited |
Definition at line 280 of file PMNS_Base.h.
Referenced by OscProb::PMNS_Base::GetAngle(), OscProb::PMNS_DensityMatrix::ProbMatrix(), OscProb::PMNS_Base::RotateH(), OscProb::PMNS_Base::RotateState(), OscProb::PMNS_Base::SetAngle(), and OscProb::PMNS_Fast::SetVacuumEigensystem().
◆ fUseCache
|
protectedinherited |
Definition at line 301 of file PMNS_Base.h.
Referenced by OscProb::PMNS_Base::FillCache(), OscProb::PMNS_Base::SetUseCache(), and OscProb::PMNS_Base::TryCache().
◆ fVcosT
|
protected |
Definition at line 201 of file PMNS_Maltoni.h.
Referenced by AlgorithmDensityMatrix(), AvgAlgo(), AvgAlgoCosT(), AvgProb(), ExtrapolationProbCosT(), and InitializeTaylorsVectors().
◆ fVInvE
|
protected |
Definition at line 191 of file PMNS_Maltoni.h.
Referenced by AlgorithmDensityMatrix(), AvgAlgo(), AvgProbMatrixLoE(), AvgProbVectorLoE(), ExtrapolationProb(), ExtrapolationProbLoE(), and InitializeTaylorsVectors().
◆ kGeV2eV
|
staticprotectedinherited |
Definition at line 217 of file PMNS_Base.h.
Referenced by AlgorithmDensityMatrix(), AvgAlgo(), AvgProbMatrixLoE(), AvgProbVectorLoE(), OscProb::PMNS_OQS::BuildDissipator(), OscProb::PMNS_OQS::BuildHVMB(), ExtrapolationProb(), ExtrapolationProbLoE(), OscProb::PMNS_Base::GetDmEff(), OscProb::PMNS_Base::GetSamplePoints(), OscProb::PMNS_Deco::PropagatePath(), OscProb::PMNS_Iter::PropagatePath(), OscProb::PMNS_Fast::SetVacuumEigensystem(), OscProb::PMNS_Decay::UpdateHam(), OscProb::PMNS_Fast::UpdateHam(), OscProb::PMNS_LIV::UpdateHam(), OscProb::PMNS_NSI::UpdateHam(), OscProb::PMNS_NUNM::UpdateHam(), OscProb::PMNS_SNSI::UpdateHam(), and OscProb::PMNS_Sterile::UpdateHam().
◆ kGf
|
staticprotectedinherited |
Definition at line 220 of file PMNS_Base.h.
Referenced by OscProb::PMNS_OQS::BuildHVMB(), OscProb::PMNS_Iter::SetExpVL(), OscProb::PMNS_Decay::UpdateHam(), OscProb::PMNS_Fast::UpdateHam(), OscProb::PMNS_LIV::UpdateHam(), OscProb::PMNS_NSI::UpdateHam(), OscProb::PMNS_NUNM::UpdateHam(), OscProb::PMNS_SNSI::UpdateHam(), and OscProb::PMNS_Sterile::UpdateHam().
◆ kK2
|
staticprotectedinherited |
Definition at line 216 of file PMNS_Base.h.
Referenced by OscProb::PMNS_OQS::BuildHVMB(), OscProb::PMNS_Iter::SetExpVL(), OscProb::PMNS_Decay::UpdateHam(), OscProb::PMNS_Fast::UpdateHam(), OscProb::PMNS_LIV::UpdateHam(), OscProb::PMNS_NSI::UpdateHam(), OscProb::PMNS_NUNM::UpdateHam(), OscProb::PMNS_SNSI::UpdateHam(), and OscProb::PMNS_Sterile::UpdateHam().
◆ kKm2eV
|
staticprotectedinherited |
Definition at line 215 of file PMNS_Base.h.
Referenced by BuildKcosT(), BuildKE(), OscProb::PMNS_Base::GetSamplePoints(), OscProb::PMNS_Base::PropagatePath(), OscProb::PMNS_Decay::PropagatePath(), OscProb::PMNS_Deco::PropagatePath(), OscProb::PMNS_Iter::PropagatePath(), OscProb::PMNS_OQS::PropagatePath(), PropagatePathTaylor(), and OscProb::PMNS_Iter::SetExpVL().
◆ kNA
|
staticprotectedinherited |
Definition at line 218 of file PMNS_Base.h.
◆ one
|
staticprotectedinherited |
Definition at line 212 of file PMNS_Base.h.
Referenced by OscProb::PMNS_Base::ResetToFlavour(), and OscProb::PMNS_DensityMatrix::ResetToFlavour().
◆ zero
|
staticprotectedinherited |
Definition at line 211 of file PMNS_Base.h.
Referenced by OscProb::PMNS_NUNM::GetAlpha(), OscProb::PMNS_LIV::GetaT(), OscProb::PMNS_LIV::GetcT(), OscProb::PMNS_NSI::GetEps(), OscProb::PMNS_Decay::PMNS_Decay(), OscProb::PMNS_Base::ResetToFlavour(), and OscProb::PMNS_DensityMatrix::ResetToFlavour().
The documentation for this class was generated from the following files:
- inc/PMNS_Maltoni.h
- src/PMNS_Maltoni.cxx